The world is currently facing an unprecedented threat from the novel coronavirus disease (COVID-19). As of June 15, 2020, 8.0 million people have been infected worldwide, more than 430,000 people have lost their lives, and no one is certain of how the situation will develop. If we hope to overcome humanity's newest foe, the world must unite to unravel and understand how it works in scientific terms. The urgent issues we now face are how to use these findings to provide accurate diagnoses, predict severity, and develop effective treatments and vaccines.
A joint research Coronavirus Task Force has been established by Keio University, Tokyo Medical and Dental University, Osaka University, The Institute of Medical Science at The University of Tokyo, the National Center for Global Health and Medicine, the Tokyo Institute of Technology, Kitasato University, and Kyoto University.
This task force comprises experts from diverse fields, including infectious diseases, virology, molecular genetics, genomic medicine, and computational science. One of the biggest threats posed by the novel coronavirus disease is that many patients with severe symptoms become critically ill within a short period of time. And saving these patients’ lives requires the allocation of vast medical resources. The task force will use state-of-the-art genomic analysis technology to reveal the genetic basis for the mechanism that causes exacerbation of COVID-19 and will work to develop an effective mucosal vaccine to protect against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Specifically, the task force will focus on the fact that the number of COVID-19 deaths per capita among Japanese people is far fewer than those in Western countries.
By comparing severe and mild/asymptomatic cases among Japanese people, the task force will search for genetic susceptibilities unique to Japanese people associated with exacerbation of COVID-19. Based on findings obtained in this way, the task force aims to develop an effective mucosal vaccine built upon proprietary technologies as well as to provide predictive modeling for COVID-19 treatment. This research is supported by the Japan Agency for Medical Research and Development.
Learn more at www.covid19-taskforce.jp (Japanese)
1. About the Coronavirus Task Force
The world is currently facing an unprecedented threat from the novel coronavirus disease (SARS-CoV-2 *1) (COVID-19 *2). According to the WHO and other infectious disease and public health experts, this pandemic will last for two to three years, and a second wave of the epidemic is expected to arrive this fall or winter. This kind of situation comes only once a century and forces us to change the way we live.
In light of the situation, we have decided to launch a university-led coronavirus task force. This project began as a discussion among a small group of doctors and researchers. But as more people die every day in Japan and around the world, our group continued to grow. Countless individuals have come together to ask how we can contribute to society through medicine and science.
The task force aims to better understand the novel coronavirus disease in specialized teams and to publish objective research and information that contributes to rational prevention and treatment. These teams will comprise those on the front line of ICU care, medical staff from university hospitals, practitioners at the forefront of community healthcare, immunologists, infectious disease experts, and even members of the general public who may not be medical experts but who are sympathetic to our cause. The mission of this task force is as follows.
In light of the situation, we have decided to launch a university-led coronavirus task force. This project began as a discussion among a small group of doctors and researchers. But as more people die every day in Japan and around the world, our group continued to grow. Countless individuals have come together to ask how we can contribute to society through medicine and science.
The task force aims to better understand the novel coronavirus disease in specialized teams and to publish objective research and information that contributes to rational prevention and treatment. These teams will comprise those on the front line of ICU care, medical staff from university hospitals, practitioners at the forefront of community healthcare, immunologists, infectious disease experts, and even members of the general public who may not be medical experts but who are sympathetic to our cause. The mission of this task force is as follows.
1) Contribute to the promotion of research, including international cooperation, through the accumulation of as many COVID-19 patient samples (DNA, RNA, plasma) as possible
2) Promote basic research that uses clinical samples
3) Promote vaccine development
As of June 15, 2020, the collection of patient samples has already begun at thirty facilities that have received approval from a research ethics committee, with 60 more facilities currently awaiting approval to start their own large-scale sample collection.
We continue to welcome the participation of as many medical institutions as possible. Our first specific research goal is to define host factors, in particular genetic factors involved in the exacerbation of COVID-19. Research findings and progress will be shared regularly on the task force’s site. We ask the citizens of Japan for their understanding and support.
We continue to welcome the participation of as many medical institutions as possible. Our first specific research goal is to define host factors, in particular genetic factors involved in the exacerbation of COVID-19. Research findings and progress will be shared regularly on the task force’s site. We ask the citizens of Japan for their understanding and support.
2. Research Background and Antiviral Immunity
COVID-19 is a significant threat to humanity. In the Western countries, in particular, the death toll from COVID-19 is already in the hundreds of thousands. While the spread of COVID-19 has become a serious social and public health issue in East Asian countries, including Japan, the fact that the mortality rate associated with the disease has remained lower than in the United states and European countries is attracting attention from around the world. Potential factors have been pointed out, including the high rate of face mask use and hand hygiene compliance, potential acquisition immunity from similar past viral outbreaks, and BCG vaccinations as well as the standards of medical care based on a universal health insurance system in Japan, in particular. But there has also been speculation that genetic differences between different populations may also play a role.
Fig. 1: COVID-19 Deaths by Country
Source: Our World in Data
Typically, when an unknown virus infects a human host, various biological mechanisms are triggered to eliminate it. These mechanisms are known collectively as immunity, of which there are two main types: innate immunity and adaptive immunity. The mechanisms of innate immunity are naturally possessed by humans and provide an early line of defense against various bacteria, viruses, and toxins. On the other hand, adaptive immunity is an immune response that comes later, where cytotoxic T cells specifically recognize and remove cells infected with a particular virus, and B cells produce specific antibodies that neutralize the virus. Once established, this immunity is “adapted” or “acquired,” and the next time the host is infected with the same virus, it can be eliminated immediately.
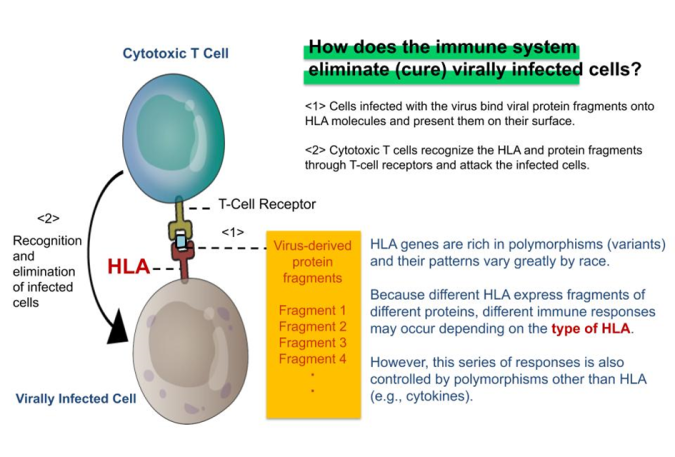
Fig. 2: HLA & Antiviral Immunity
Immune response to a virus also varies from individual to individual due to differences in the nucleotide sequences of genes called “polymorphisms.” This variability means that even if two people become infected with the same virus, polymorphic variations may yield different immune responses. In an immune response, a group of molecules collectively referred to as HLA (*3) acts as a command tower that induces adaptive immunity, specifically binding to bits of virus proteins, generally referred to as epitopes. The recognition by T-cell receptors of epitopes bound to HLA is the key to the function of adaptive immunity, which is expressed in the function of cytotoxic T cells and antibody-producing B cells. There are many types of HLA genes (HLA-A, B, C, DR, DQ, DQ, DP, etc.) and the polymorphism in each HLA (e.g., HLA-A) are known to vary for individual to individual. This is thought to be related not only to differences in responsiveness to viruses but also to susceptibility to seasonal allergies and many other immune-mediated diseases. On the other hand, these immune responses, including innate immunity, are also strongly affected by individual variation other than polymorphic HLA genes, suggesting that various polymorphisms, in addition to HLA, play a significant role in the exacerbation of COVID-19.
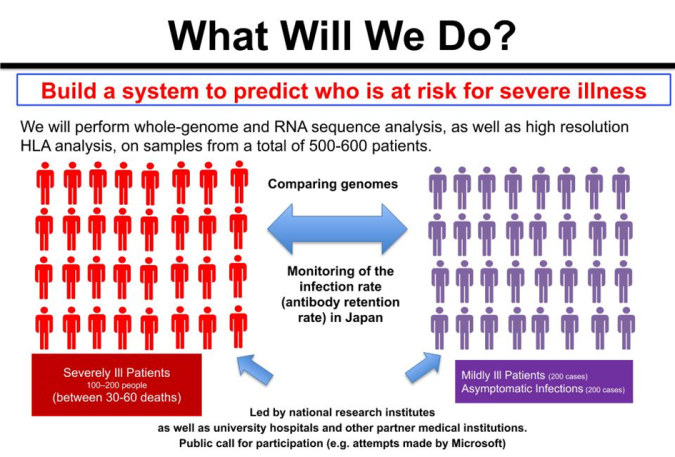
Fig. 3: Finding Genetic Polymorphisms that Influence COVID-19 Exacerbation
We ask, "Are polymorphisms, including HLA, involved in the exacerbation of COVID-19?” And if so, then we must also ask, "What kinds of polymorphism in which genes are involved?" If such polymorphism is present, we can presume that the frequency will differ between severe and mild or asymptomatic cases.
Therefore, our research aims to determine all gene sequences for patients who have become severely ill with COVID-19 as well as patients who had only mild symptoms or were asymptomatic and to find polymorphisms that differ in frequency between these two groups. One factor involved in the exacerbation of COVID-19 that has drawn attention is the phenomenon known as a cytokine (*4) storm. During these storms, proteins called "cytokines," which regulate the function of immune cells and the body’s immune response, are generated in excess, resulting in damage not only to virally infected cells but also to healthy cells and organs. Such a phenomenon may also be affected by genetic polymorphism.
Meanwhile, the development of an effective vaccine is the key to overcoming emerging viral diseases. Identifying genes involved in exacerbation of COVID-19 that are unique to Japanese people would be useful not only for predicting that exacerbation and developing effective therapeutic agents but would accelerate vaccine development. The research team also possesses unique technologies such as molecular needle and organoid (*5) technology. These proprietary technologies are expected to produce synergistic effects in the development of a SARS-CoV-2 vaccine.
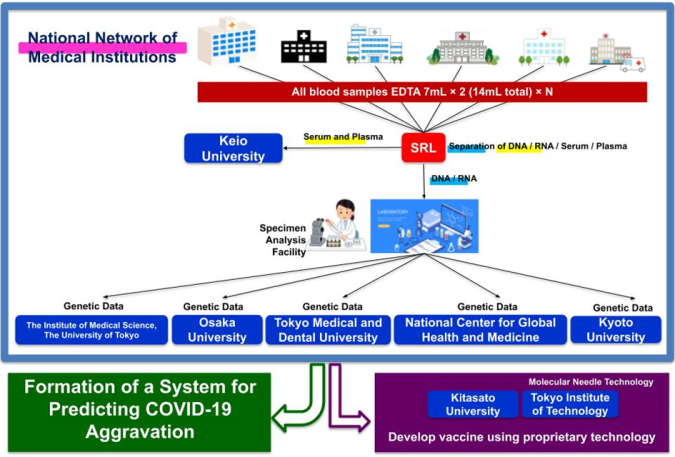
Fig. 4: Coronavirus Task Force Overview
3. Research Planning
The Coronavirus Task Force research team plans to collect 600 blood samples from Japanese COVID-19 patients from our joint research institutes (95 hospitals as of June 15). Using these samples, we will conduct comprehensive analyses such as high-resolution HLA analysis (*6), SNP array analysis (*7), whole-genome sequence analysis (*8), and T-cell repertoire analysis. By comparing severe cases with mild and asymptomatic cases, we aim to identify the genes involved in the exacerbation of COVID-19 in Japanese patients.Determining the target epitope for a vaccine is a critical step in vaccine development, but epitope prediction has proved to be a challenge. After identifying the genes involved in the exacerbation of COVID-19 in Japanese patients, the Coronavirus Task Force research team will aim to identify candidate antigens for SARS-CoV-2 using supercomputer simulations and will develop a vaccine better suited for preventing the exacerbation of the disease.
A series of genomic analyses have already been approved by ethics committees at Keio University and more than 30 medical facilities, and we have begun sequential sampling from authorized medical institutions while continuing to reach out to more medical institutions to cooperate.
Using proprietary molecular needle technology, the Coronavirus Task Force also aims to develop an effective mucosal vaccine (*9) for SARS-CoV-2. This vaccine will be based on information gained from genetic and immunological analyses of severe and mild or asymptomatic COVID-19 cases.
The molecular needle is a technology that transports protein molecules using a protein needle (molecular needle) approximately 10 nanometers in length at the end of the tail of a bacteriophage. Molecular needles can spontaneously translocate through a cell membrane and enter a cell, which makes it possible to deliver viral antigens to the body (inside of cells) to efficiently induce an immune response to a virus. To do this, we will take the proteins and peptide sequences of the viruses that comprise the vaccine and fuse them to the needle. Inoculations will then be administered intranasally via the mucous membranes of the nasal cavity or sublingually. Though conventional vaccines are mostly administered through subcutaneous injection, molecular needle technology can be administered by being inhaled through the nose, dripped under the tongue, or swallowed as a capsule, which means it is relatively safe and painless.
In this study, we will search for the genes involved in the exacerbation of COVID-19, but will also comprehensively examine the genes associated with COVID-19 immune responses. It is expected that the results will be used to effectively find a portion of the protein sequence of SARS-CoV-2—the virus that causes COVID-19—and will effectively prevent exacerbation of COVID-19 and help inform prevention and healing. We plan to develop a molecular needle vaccine that fuses the SARS-CoV-2 proteins and peptide sequences discovered in this way.
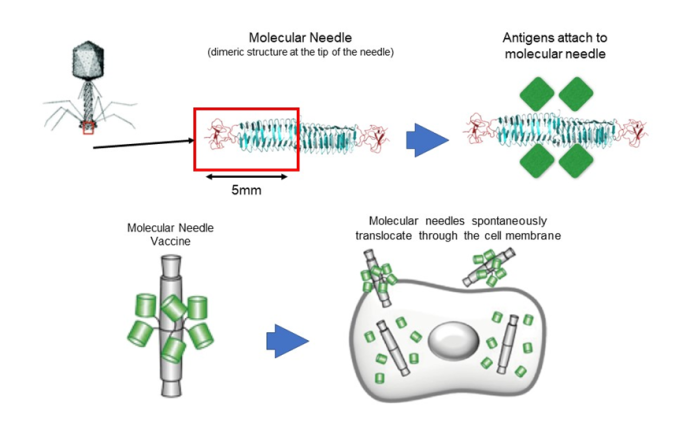
Fig. 5: Development of an Adjuvant-Free COVID-19 Vaccine for Mucosal Immunity
4. Future Developments
The Coronavirus Task Force is currently seeking joint research facilities. (https://www.covid19-taskforce.jp/) We plan to analyze the human genes of samples collected in July of this year and quickly compile the results of our research by September. We will reveal, through analysis, the genes associated with the exacerbation of COVID-19 among Japanese people in order to predict exacerbation at the time of diagnosis. Using these genes as an index for medical administration and for informing medical guidelines is expected to help prevent medical collapse during a potential second or third wave of COVID-19. Based on the immunological genetic information obtained, it is also expected to inform the design of a SARS-CoV-2 vaccine that will be adapted for many Japanese people.5. Support
This research is a Drug Discovery Support Promotion Project of the Japan Agency for Medical Research and Development (AMED): Issues in Vaccine Research and Development Regarding the Novel Coronavirus Disease (COVID-19) “Development of a Molecular Needle Mucosal COVID-19 Vaccine Based on Genetic Information Regarding the Novel Coronavirus Disease” (Principal Investigator: Takanori Kanai)This research will be conducted based on AMED research funds and contributions from donors who have supported this project.
6. Task Force Members
Keio University: Prof. Takanori Kanai, Prof. Koichi Fukunaga, Prof. Naoki Hasegawa, Professor Toshiro Sato, Hospital Director General Yuko KitagawaTokyo Medical and Dental University: M&D Data Science Center Director Satoru Miyano, Prof. Ryuji Koike, Prof. Masumi Ai, and Executive Director, Vice President Akinori Kimura
Osaka University: Prof. Yukinori Okada, Prof. Atsushi Kumanogoh (Graduate School of Medicine)
The Institute of Medical Science, The University of Tokyo: Prof. Seiya Imoto
National Center for Global Health and Medicine: Director Katsushi Tokunaga
Tokyo Institute of Technology: Prof. Takashi Ueno (School of Life Science and Technology)
Kitasato University: Prof. Kazuhiko Katayama, Prof. Tomomi Takano
Kyoto University: Prof. Seishi Ogawa
Research Notes
1 Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)The strain of coronavirus that causes the novel coronavirus disease (COVID-19). Named SARS-CoV-2 because of its similarity to the SARS coronavirus that emerged in 2002.
2 COVID-19
An abbreviation of coronavirus disease 2019, a new strain of coronavirus discovered in 2019. It refers to the disease caused by the novel coronavirus. By the end of 2019, an infection that had started in Wuhan, China had spread across the world. Those infected with the novel coronavirus develop symptoms including fever, cough, and shortness of breath. When the infection spreads to the lungs, it causes pneumonia, which can lead to respiratory compromise.
3 HLA
Just as there are different types of red blood cells such as A, B, AB, and O, there are different types of cells all throughout the body, including white blood cells. These types are called human leukocyte antigens (HLA). HLA types are highly diverse and vary significantly among individuals, but they play a key role in immune responsiveness to antigens. For this reason, it is known that each specific HLA type has different immune responsiveness to specific antigens.
4 Cytokine
A general term for proteins secreted by cells, which play a significant role in signaling between cells, especially in immunity and inflammation. Many studies have reported that infections caused by SARS-CoV-2 also cause abnormalities in immunity and inflammation.
5 Organoid
Cultivated in the form of sheets using conventional cell culture technology. The term organoid refers to cultured cells grown as a three-dimensional structure using hydrogels as scaffolding and nutrients known as growth factors. From a single stem cell, it is possible to create a tissue-like structure in vivo and infinitely increase the number of healthy cells found in various tissues such as the respiratory tract and lungs.
6 High-Resolution HLA Analysis
The HLA gene region is highly diverse, and its sequence differs among populations. In this research, we aim to perform detailed HLA analysis using a next-generation sequencer.
7 SNP Array Analysis
A single-nucleotide polymorphism (SNP, pronounced snip) is a single base-pair difference in the human genome and can make a difference in individuals' susceptibility to illness. An SNP array is the comprehensive analysis of SNPs across a person’s entire genome.
8 Whole-Genome Sequence Analysis
Analysis of the entire genomic DNA sequence. A large number of genes are examined simultaneously with a next-generation sequencer that can read large amounts of genomic data at high speeds.
9 Mucosal Vaccine
A vaccine administered orally or nasally to mucous membranes, targeting mucosal surfaces such as the intestines and nasopharynx. Compared with injectable vaccines, which are currently more commonly used, mucosal vaccines induce both systemic and mucosal immune responses. They are expected to be used against pathogens that enter through mucosal surfaces such as the respiratory tract and digestive system. Their practicality is also expected to lead to increased use as a less invasive alternative to subcutaneous vaccines.