| Key Points of Presentation |
|---|
|
Presentation Outline
As the novel coronavirus keeps mutating during the pandemic, strains with higher infectivity or increased vaccine-resistance strains threaten to emerge. It is extremely important to monitor strains and to identify emerging strains in order to implement preventive measures. Further, the mutation pattern of a viral isolate is important to identify possible parental strains and transmission routes. With transmission route analysis, high risk places and activities will be revealed, enabling authorities to implement effective preventive measures. As of May 2021, more than 1.7 million genome sequences of SARS-CoV-2 have been released globally. To utilize such information, the Human Genome Center, the Institute of Medical Science, The University of Tokyo and IBM have developed the HGC SARS-CoV-2 Variant Browser on the supercomputer SHIROKANE. The system helps to monitor strains, to detect emerging variants of concern, and to identify infection routes.
Details of Presentation
Background /Challenge
The worldwide pandemic of COVID-19 continues, and Japan remains in a difficult situation. The coronavirus has accumulated mutations amid the prolonged pandemic. Occasionally, mutations induce novel characteristics, impacting on infectious disease control measures.
On the other hand, by utilizing mutation data, we can potentially acquire useful insights such as "what kind of mutations the novel coronavirus strain harbors," "when and from which country the strain arrived", and "how the strain spread.” The mutation data becomes crucial information to learn about the expansion status of existing mutant strains and detect emerging strains. To that end, the genome data should be quickly analyzed. For example, as of May, 2021, more than 1.7 million novel coronavirus sequences have been registered in the Global Initiative on Sharing Avian Influenza Data (GISAID)*1. However, a system for rapidly analyzing a large number of genomes has been lacking was needed.
Especially for mass gathering events such as sports, concerts and festivals, preventive measures are becoming more important to mitigate the risk of COVID-19 spreading. Verification studies are underway to evaluate the risks of mass gathering events in the United Kingdom*2 and the Netherlands*3. Also, in Japan, such risk evaluations have been conducted in professional sports. Pre-events of the Tokyo Olympics and Paralympics have begun. If the Olympic and Paralympic games are held, higher domestic mobility is expected in Japan, accompanied by the arrival of athletes and staff from overseas. In addition, more people will resume social activities as they are vaccinated. These factors will contribute to the emergence of immune escape variants. Therefore, it is very important to monitor strains. Furthermore, mutation data will enable us to figure out infection routes, which could lead to the identification of high-risk places and activities.
The Human Genome Center of the Institute of Medical Science, The University of Tokyo and IBM have developed the HGC SARS-CoV-2 Variant Browser which can monitor novel coronavirus strains and figure out the infection routes, thus establishing a system that can tackle the above-mentioned bottleneck challenges.
Collaboration Overview
The HGC SARS-CoV-2 Variant Browser was developed based on the technologies of SARS-CoV-2 Variant Annotator*4 and the SARS-CoV-2 Variant Browser*5 developed by IBM Research and IBM Garage for COVID-19. The browser is in operation at Human Genome Center. In particular, in collaboration with IBM, we have deployed a new feature to figure out the origins and timings of the introduction of exogenous strains and their infection routes in Japan. Timed with this, Human Genome Center has also released the data analysis pipeline HGC_CovidPipeLine*6 for analyzing raw sequencing data of the novel coronavirus so that it can be visualized with the HGC SARS-CoV-2 Variant Browser.
The HGC SARS-CoV-2 Variant Browser enables us to perform the following analyses.
< Estimating the origins and timing of exogenous strain arrivals and their infection routes>
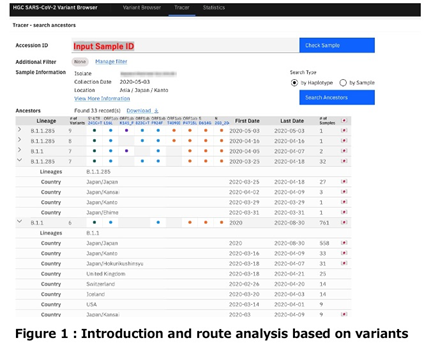
This enables us to figure out when and where the novel coronavirus is introduced into Japan from genomic fingerprints of existing strains. In addition, it also enables us to trace in which area the strain is detected, so we can examine whether it stays where it is or spreads to other areas (Figure 1).
< Monitor the novel coronavirus strains>
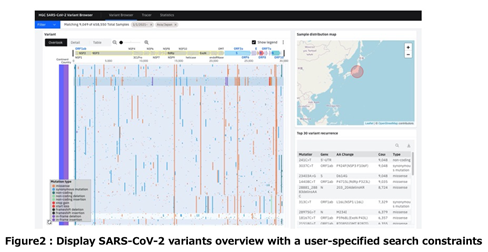
When a new mutation is identified in Japan, this enables us to search whether it has been reported elsewhere. In addition, we can perform spatiotemporal searches to monitor strains using user-specified search constraints (Figure 2).
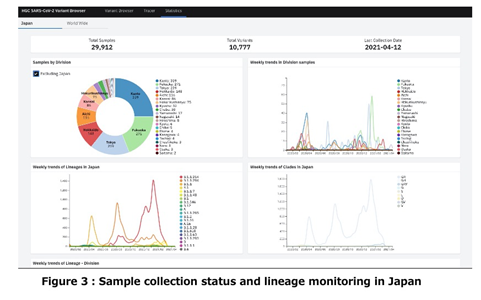
The accumulated novel coronavirus genomes are important for the analysis. Meanwhile, SARS-CoV-2 genome data availability differs by prefecture in Japan, as well as by country in the world. In order to understand such data characteristics, we have implemented a viewer to monitor the number of registered genomes released in the world and Japan together with their lineages (Figure 3).
Since the novel coronavirus strains will continue to accumulate mutations, there are concerns about the emergence of vaccine-resistance strains and reverse-zoonosis strains. Throughout our initiatives, we continue to tackle these issues. Additionally, in order to promptly obtain information on variants of concern, we will establish a system that analyzes genomic data in collaboration with various organizations and municipal governments.
The Human Genome Center of the Institute of Medical Science, The University of Tokyo has been working hard to conduct research to analyze SARS-CoV-2 genomes and bring new treatment methods, leveraging expertise obtained through genome and data science researchers. By utilizing this system for analysis of the novel coronavirus genome collected by the COVID-19 task force and monitoring of virus transmission caused by increased mobility during the Olympic and Paralympic games, we will make efforts to contribute to basic research on COVID-19 and formulation of preventive measures.
IBM has been taking various initiatives related to COVID-19. The Variant Annotator and the Variant Browser for visualization provided to The Institute of Medical Science, The University of Tokyo, are part of IBM's research and development efforts since the early stages of the COVID-19 pandemic. Additionally, IBM analyzed SARS-CoV-2 genomes in Japan*7. By harnessing its IT technologies, IBM plans to continue to tackle COVID-19 issues.
Inquiries
Research contactHuman Genome Center, The Institute of Medical Science, The University of Tokyo
Seiya Imoto Director / Professor
URL: https://www.ims.u-tokyo.ac.jp/imsut/en/index.html
Media Contacts
The Institute of Medical Science, The University of Tokyo
URL:https://www.ims.u-tokyo.ac.jp/imsut/en/index.html
IBM Japan Communications
URL:https://www.ibm.com/jp-ja
Reference
*1 GISAID: https://www.gisaid.org/*2 The United Kingdom Government. Information on the Events Research Programme. 2021. https://www.gov.uk/government/publications/guidance-about-the-events-research-programme-erp-paving-the-way-for-larger-audiences-to-attend-sport-theatre-and-gigs-safely-this-summer/guidance-on-the-events-research-programme
*3 de Vrieze J. Dutch studies bring back the fun—but are they good science? Science 2021; 372(6541): 447
*4 Variant Annotator: Koyama T., Platt D., Parida L. “Variant analysis of SARS-CoV-2 genomes.” Bull World Health Organ. 2020;98:495-504.
*5 SARS CoV-2 Variant Browser - IBM Functional Genomics Platform: https://ibm.biz/functional-genomics
*6 HGC_CovidPipeLine:https://www.hgc.jp/~ppl_ht/CovidPipeLine/latest/docs/
*7 Tokumasu R., Weeraratne D., Snowdon J., Parida L., Kudo M., Koyama T. “Introductions and evolutions of SARS-CoV-2 strains in Japan” medRxiv 2021.02.26.21252555.