Comprehensive identification of viral proteins encoded by viral genes is required to understand the pathophysiology of viral infections. A research team led by Professor Yasushi Kawaguchi of the Institute of Medical Science, the University of Tokyo, conducted mass spectrometry specialized for novel synthetic proteins of viruses, and developed a new decoding method for viral genes that can easily and quickly obtain even non-canonical genetic information.
Using this new decoding method, they identified nine novel proteins encoded by herpes simplex virus type 1(HSV-1) (*1) and found that one of them, piUL49, is a pathogenic factor that specifically controls the onset of herpes encephalitis (*2).
These results were published in ”Nature Communications” on September 29, 2020.
Protein piUL49 is involved in brain-specific viral proliferation and the onset mechanism of viral encephalitis
It is difficult to decipher the whole picture of diverse and complex genetic information hidden in the viral genome with conventional technology, and the development of a new method has been required for decoding viral genes, especially those encoding non-canonical translational elements.Many viruses are known to shut-off new synthesis of host proteins. Focusing on this property, the research team purified newly synthesized proteins by the BONCAT method (*3) and performed high-sensitivity mass spectrometry.
They found that most of the peptides obtained were derived from HSV-1, including peptides from viral proteins encoded by nine novel HSV-1 genes. All the newly identified HSV-1 genes encode non-canonical translational products.
They named one of these novel viral proteins piUL49. Using analysis of a mouse model of HSV-1 infection, they clarified that piUL49 is involved in brain-specific viral proliferation and the onset mechanism of viral encephalitis. For details of the research, please see the paper.
Expected to lead to the development of new treatments for HSV encephalitis
The entire base sequence of the HSV-1 genome was determined about 20 years ago, and it is thought that the decoding of the viral genes encoding canonical translational elements has already been completed. However, information on the decoding of viral genes encoding non-canonical translational elements has been limited.
It is of great academic significance to discover nearly 10 new HSV genes and to clarify that one of them encodes piUL49, which is involved in the development of viral encephalitis.
Professor Kawaguchi, the lead scientist of this research, stressed the importance of their finding as follows. "Elucidation of the onset mechanism of encephalitis through piUL49 will greatly contribute to the understanding of the high central nervous system orientation of HSV-1. We hope that the results will lead to the development of new treatments for HSV-1 encephalitis.
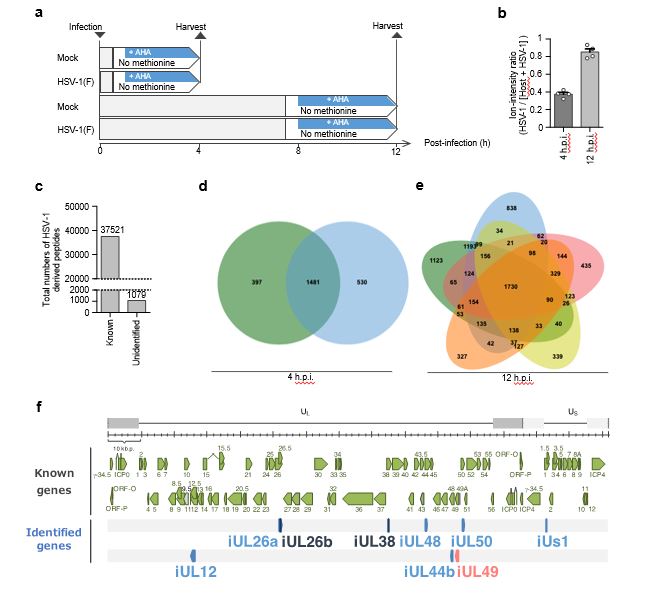
Fig. 1: Identification of previously unidentified HSV-1 CDSs by chemical proteomics.
This diagram illustrates the procedure of AHA labeling experiments for BONCAT-based MS-analyses (n = 2). In each of the two experiments, MS-analyses were performed twice. b Ion-intensity of viral polypeptides relative to that of viral and host polypeptides. Each value is the mean ± SEM of four MS-analyses. c Total numbers of peptides detected in HSV-1-infected cells at 4 h and 12 h post-infection and mapped to known or previously unidentified HSV-1 Coding Sequences (CDSs) in the experiments shown in Fig. 1a and Supplementary Fig. 1a. The number is shown above the bars. d Venn diagrams showing the results of two independent AHA-labeling experiments by 4 h post-infection using wild-type HSV-1(F)-infected cells as described in Fig. 1a. The numbers indicate the number of peptides derived from HSV-1 CDSs. e Venn diagrams showing the results of five independent AHA-labeling experiments by 12 h post-infection using wild-type HSV-1(F) infected cells as described in Fig. 1a (n = 2) and Supplementary Fig. 1a (n = 3). The numbers indicate the number of peptides derived from HSV-1 CDSs. f Schematic diagram of the genome of wild-type HSV-1 and the map of HSV-1 CDSs known and identified in this study.
It is of great academic significance to discover nearly 10 new HSV genes and to clarify that one of them encodes piUL49, which is involved in the development of viral encephalitis.
Professor Kawaguchi, the lead scientist of this research, stressed the importance of their finding as follows. "Elucidation of the onset mechanism of encephalitis through piUL49 will greatly contribute to the understanding of the high central nervous system orientation of HSV-1. We hope that the results will lead to the development of new treatments for HSV-1 encephalitis.

Fig. 1: Identification of previously unidentified HSV-1 CDSs by chemical proteomics.
This diagram illustrates the procedure of AHA labeling experiments for BONCAT-based MS-analyses (n = 2). In each of the two experiments, MS-analyses were performed twice. b Ion-intensity of viral polypeptides relative to that of viral and host polypeptides. Each value is the mean ± SEM of four MS-analyses. c Total numbers of peptides detected in HSV-1-infected cells at 4 h and 12 h post-infection and mapped to known or previously unidentified HSV-1 Coding Sequences (CDSs) in the experiments shown in Fig. 1a and Supplementary Fig. 1a. The number is shown above the bars. d Venn diagrams showing the results of two independent AHA-labeling experiments by 4 h post-infection using wild-type HSV-1(F)-infected cells as described in Fig. 1a. The numbers indicate the number of peptides derived from HSV-1 CDSs. e Venn diagrams showing the results of five independent AHA-labeling experiments by 12 h post-infection using wild-type HSV-1(F) infected cells as described in Fig. 1a (n = 2) and Supplementary Fig. 1a (n = 3). The numbers indicate the number of peptides derived from HSV-1 CDSs. f Schematic diagram of the genome of wild-type HSV-1 and the map of HSV-1 CDSs known and identified in this study.
Research Notes
(*1) Herpes simplex virus type 1 (HSV-1)
Herpes simplex virus type 1 (HSV-1) is a virus that causes a variety of human diseases including encephalitis, keratitis, and mucocutaneous and skin diseases such as herpes labialis, genital herpes, and herpetic whitlow.
(*2) Herpes encephalitis
Herpes encephalitis is caused by an infection of the brain with HSV. The case fatality rate without treatment is as high as 70% or more. Even with antivirals, 10-15% of patients die. Problems such as serious sequelae have also been pointed out.
(*3) BONCAT method
BONCAT (BioOrthogonal Non-Canonical Amino acid Tagging) is a tool for tracking protein synthesis on the level of single cells within communities and whole organisms. A method that can concentrate only newly synthesized proteins. This method was first reported in 2006 by Dr. DC. Daniela of the University of California et al.
Herpes simplex virus type 1 (HSV-1) is a virus that causes a variety of human diseases including encephalitis, keratitis, and mucocutaneous and skin diseases such as herpes labialis, genital herpes, and herpetic whitlow.
(*2) Herpes encephalitis
Herpes encephalitis is caused by an infection of the brain with HSV. The case fatality rate without treatment is as high as 70% or more. Even with antivirals, 10-15% of patients die. Problems such as serious sequelae have also been pointed out.
(*3) BONCAT method
BONCAT (BioOrthogonal Non-Canonical Amino acid Tagging) is a tool for tracking protein synthesis on the level of single cells within communities and whole organisms. A method that can concentrate only newly synthesized proteins. This method was first reported in 2006 by Dr. DC. Daniela of the University of California et al.
Funding
This study was supported by Grants for Scientific Research and Grant-in-Aid for Scientific Research (S) (20H05692) from the Japan Society for the Promotion of Science (JSPS), Grants for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports and Technology of Japan (16H06433, 16H06429, 16H06430, 16K21723, 17H05610, 18H04968, 20H04853, and 24115007), grants from the Japan Agency for Medical Research and Development (AMED) through the Program of Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) (JP18fm0108006), the Research Program on Emerging and re-emerging Infectious Diseases (19fk018105h0001), and the Japan Program for Infectious Diseases Research and Infrastructure (20wm0125002h0001, 20wm0225017s, 20wm0225009h), the grant for International Joint Research Project of the Institute of Medical Science, the University of Tokyo, grants from the Takeda Science Foundation, the Naito Foundation, and the Ichiro Kanehara Foundation, the Kanae Foundation for the Promotion of Medical Science, the Waksman Foundation of Japan, and the GSK Japan Research Grant 2018. The computational resource was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, and The University of Tokyo.
About the research
1) Journal Article
Akihisa Kato, Shungo Adachi, Shuichi Kawano, Kousuke Takeshima, Mizuki Watanabe, Shinobu Kitazume, Ryota Sato, Hideo Kusano, Naoto Koyanagi, Yuhei Maruzuru, Jun Arii, Tomohisa Hatta, Tohru Natsume & Yasushi Kawaguchi. “Identification of a herpes simplex virus 1 gene encoding neurovirulence factor by chemical proteomics.” Nat Commun 11, 4894 (2020).
DOI:10.1038/s41467-020-18718-9
URL : https://www.nature.com/articles/s41467-020-18718-9
DOI:10.1038/s41467-020-18718-9
URL : https://www.nature.com/articles/s41467-020-18718-9
2) Publication Journal
Nature Communications
https://www.nature.com/ncomms/
https://www.nature.com/ncomms/