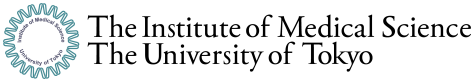A new diagnostic method for COVID-19, SARS-CoV-2, using the Japanese genome editing technology CRISPR-Cas3 (*1) has been developed that can accurately detect COVID-19 in 40 minutes. A research team working in The Institute of Medical Science, The University of Tokyo, published their method on iScience on January 30, 2022.
The new diagnostic method, named "CONAN (*2)," was the first to use CRISPR-Cas3 to detect the new coronavirus (SARS-CoV-2). The test is performed using a nasopharyngeal swab to detect viral RNA.
CRISPR-Cas3 is a genome-editing tool developed in Japan that artificially cuts DNA via a multiprotein complex. It was developed in 2019 by the research team leader, Professor Tomoji Mashimo, The Institute of Medical Science, The University of Tokyo, and other researchers.
CRISPR-Cas-3 is a Class 1 CRISPR system that can be programmed to target a defined gene sequence. The team used this mechanism to detect trace amounts of viral RNA found in genomic samples of patients infected with SARS-CoV-2.
The research team found a positive concordance rate (PPA) of 90% (9/10 cases) and a negative concordance rate (NPA) of 95.3% (20/21 cases), which rivals the PCR test method in detection sensitivity and specificity. In addition, the team claimed success in detecting the presence or absence of viral RNA down to the tens-of-copies level with a test strip in only 40 minutes.
Please see the iScience for more details.
Speed, simplicity, adaptability, and low cost
To summarize the results of this study, the CONAN method has the following advantages over PCR and antigen test methods.
1) The virus can be detected within 40 minutes from patient-derived samples.
2) Only general reagents, test papers, and constant temperature heat blocks are required for testing.
3) The method can detect as few as several dozen viral RNA copies with high sensitivity and accuracy.
4) Since CRISPR-Cas3 can detect even a single base difference, the method can be readily adapted for detecting new circulating mutations in the virus found to confer drug resistance or increased virulence.
5) The method is not limited to SARS-CoV-2; the nucleotide sequences of any virus can be detected.
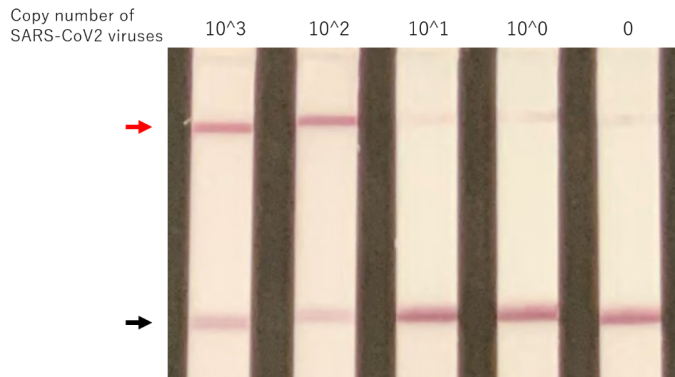
In a nutshell, by utilizing CONAN, the new COVID-19 diagnostic method can be said to combine the advantages of PCR-based methods in sensitivity and accuracy with those of antigen testing methods in speed, simplicity, and low cost (Figure 1).
1) The virus can be detected within 40 minutes from patient-derived samples.
2) Only general reagents, test papers, and constant temperature heat blocks are required for testing.
3) The method can detect as few as several dozen viral RNA copies with high sensitivity and accuracy.
4) Since CRISPR-Cas3 can detect even a single base difference, the method can be readily adapted for detecting new circulating mutations in the virus found to confer drug resistance or increased virulence.
5) The method is not limited to SARS-CoV-2; the nucleotide sequences of any virus can be detected.
In a nutshell, by utilizing CONAN, the new COVID-19 diagnostic method can be said to combine the advantages of PCR-based methods in sensitivity and accuracy with those of antigen testing methods in speed, simplicity, and low cost (Figure 1).

Figure 1: Detection of new coronavirus RNA by lateral flow test paper (red arrow: positive)
Figure 2: Rapid diagnostic tests against infectious diseases
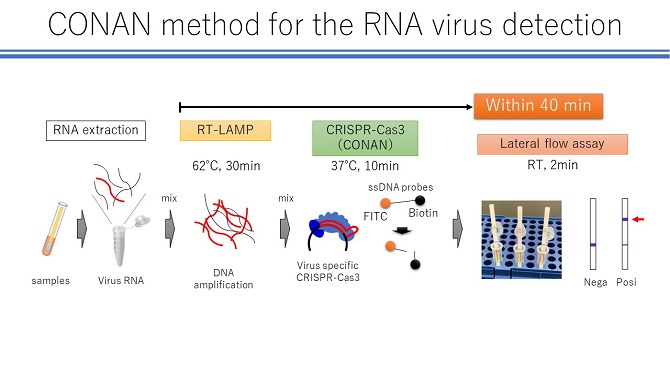
Figure 3: CONAN method for the RNA virus detection

Figure 2: Rapid diagnostic tests against infectious diseases

Figure 3: CONAN method for the RNA virus detection
Applicable to influenza diagnosis
In the future, the research team will make a kit through C4U (http://www.crispr4u.jp/en/), a Japanese university-based bio-venture company. The team intends to put the method into practical use as a COVID-19 rapid diagnostic agent that can be easily used in the medical field.
Professor Mashimo says: "If CONAN is put to practical use as a COVID-19 diagnostic method and used in various places such as medical facilities, airports, and outdoors, it will greatly contribute to the prevention of the further spread of the new coronavirus."
According to the research team, CONAN also succeeded in detecting viral RNAs of type A (H1N1pdm09, H3N2), which account for more than 95% of influenza circulating every year, using the same method.
The research team has declared that they will proceed with the development of a new influenza diagnostic at the same time.
Professor Mashimo says: "If CONAN is put to practical use as a COVID-19 diagnostic method and used in various places such as medical facilities, airports, and outdoors, it will greatly contribute to the prevention of the further spread of the new coronavirus."
According to the research team, CONAN also succeeded in detecting viral RNAs of type A (H1N1pdm09, H3N2), which account for more than 95% of influenza circulating every year, using the same method.
The research team has declared that they will proceed with the development of a new influenza diagnostic at the same time.
Research Notes
(*1) CRISPR-Cas3
Many bacteria have a defense system called "CRISPR-Cas system" that resembles adaptive immunity. CRISPR-Cas3 is a CRISPR system belonging to Class 1 among CRISPR systems of bacteria and archaea. It was developed in 2019 as a domestic genome editing tool that artificially cuts DNA with a multiprotein complex.
(*2) CONAN
Cas3 Operated Nucleic Acid detectioN method. By using a mixture of CRISPR-Cas3 protein that recognizes the target sequence and a DNA probe for detection as a biosensor, it is possible to detect quickly and accurately whether or not the target sequence is present in a sample.
Many bacteria have a defense system called "CRISPR-Cas system" that resembles adaptive immunity. CRISPR-Cas3 is a CRISPR system belonging to Class 1 among CRISPR systems of bacteria and archaea. It was developed in 2019 as a domestic genome editing tool that artificially cuts DNA with a multiprotein complex.
(*2) CONAN
Cas3 Operated Nucleic Acid detectioN method. By using a mixture of CRISPR-Cas3 protein that recognizes the target sequence and a DNA probe for detection as a biosensor, it is possible to detect quickly and accurately whether or not the target sequence is present in a sample.
About the research
Journal name: iScience
Kazuto Yoshimi, Kohei Takeshita, Seiya Yamayoshi, Satomi Shibumura, Yuko Yamauchi, Masaki Yamamoto, Hiroshi Yotsuyanagi, Yoshihiro Kawaoka, Tomoji Mashimo “ “Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3” iScience: January 30, 2022.
DOI:10.1016/j.isci.2022.103830
Kazuto Yoshimi, Kohei Takeshita, Seiya Yamayoshi, Satomi Shibumura, Yuko Yamauchi, Masaki Yamamoto, Hiroshi Yotsuyanagi, Yoshihiro Kawaoka, Tomoji Mashimo “ “Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3” iScience: January 30, 2022.
DOI:10.1016/j.isci.2022.103830
Journal name: Pre-print server "medRxiv" (published on June 2, 2020)
Kazuto Yoshimi, Kohei Takeshita, Seiya Yamayoshi, Satomi Shibumura, Yuko Yamauchi, Masaki Yamamoto, Hiroshi Yotsuyanagi, Yoshihiro Kawaoka, Tomoji Mashimo “Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3” medRxiv : June 2.
DOI: 10.1101/2020.06.02.20119875
URL: https://www.medrxiv.org/content/10.1101/2020.06.02.20119875v1
Funding
This project was supported in part by JSPS KAKENHI Grant Number 18H03974 (T.M.) and 19K16025 (K.Y.). Partially support also came from the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP20am0101070 (support numbers 1251 and 2463), from Research Program on Emerging and Re-emerging Infectious Diseases from AMED (19fk0108151, JP19fk0108113), and from the National Institutes of Allergy and Infectious Diseases funded Center for Research on Influenza Pathogenesis (CRIP; HHSN272201400008C).