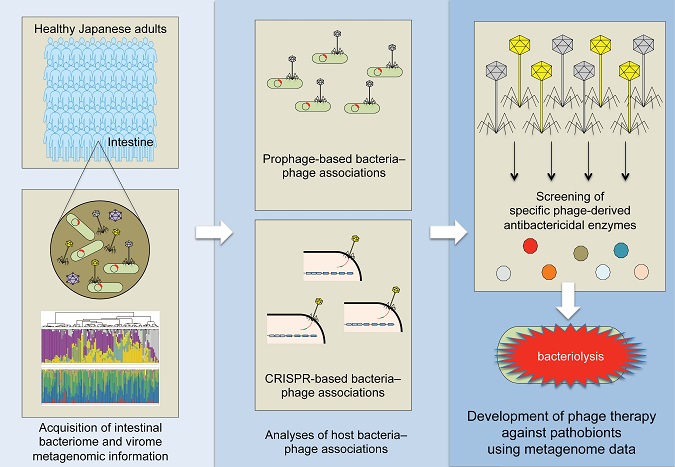
In a pioneer study published in Cell Host & Microbe –Researchers at Osaka City University and The Institute of Medical Science, The University of Tokyo, reported intestinal bacterial and viral metagenome information from the fecal samples of 101 healthy Japanese individuals. This analysis, leveraging host bacteria–phage associations, detected phage-derived antibacterial enzymes that control pathobionts. As proof-of-concept, phage-derived endolysins are shown to regulate C. difficile infection in mice.
Abnormalities in human intestinal microflora, known as dysbiosis, are connected to various diseases. Altered microbial diversity impairs the beneficial effects of host intestinal microflora, which cause some symbiotic commensal bacteria to acquire virulence traits, proliferate, and become directly involved in the development of disease. These bacteria are referred to as “pathobionts”, which are distinct from opportunistic pathogens.
C. difficile, which is a Gram-positive, spore-forming anaerobic bacterium, is a pathobiont and the representative cause of nosocomial diarrhea following antibiotic treatment. Since antibiotic usage has the risk of killing beneficial bacteria and promoting dysbiosis, the development of methods to specifically manipulate intestinal pathobionts is essential.
“Phages were sure to be applicable as a highly specific therapy for intestinal pathobiont elimination”, believed Professor Satoshi Uematsu. The infectious associations between phages and bacteria in the human intestine is essential information for the development of phage therapies. Known as “viral dark matter” as they had yet to be understood, researchers obtained metagenome information about bacteria-phage associations from the fecal samples of 101 healthy individuals through the development of a virome analysis pipeline. Based on this information, researchers screened C. difficile-specific phages and identified novel antibacterial enzymes, both in vitro and in vivo.
“The accumulation of more metagenomic information on intestinal phages and bacteria will open up the possibility of developing treatments for a variety of dysbiosis-related diseases”, say Dr. Kosuke Fujimoto and Prof. Seiya Imoto.
About the research
Kosuke Fujimoto1,2,3, Yasumasa Kimura4, Masaki Shimohigoshi1, Takeshi Satoh4, Shintaro Sato1,5, Georg Tremmel6, Miho Uematsu1, Yunosuke Kawaguchi1, Yuki Usui4, Yoshiko Nakano1, Tetsuya Hayashi1, Koji Kashima7, Yoshikazu Yuki7, Kiyoshi Yamaguchi8, Yoichi Furukawa8, Masanori Kakuta6, Yutaka Akiyama9, Rui Yamaguchi6, Sheila E. Crowe10, Peter B. Ernst11,12,13, Satoru Miyano6, Hiroshi Kiyono11,12,14,15, Seiya Imoto16,17*, Satoshi Uematsu1,2,3,17,18* (*corresponding author), 11 July 2020, "Metagenome Data on Intestinal Phage–Bacteria Associations Aids the Development of Phage Therapy Against Pathobionts"Cell Host & Microbe. (IF=15.923)
1Department of Immunology and Genomics, Osaka City University Graduate School of Medicine, Osaka 545-8585, Japan
2Division of Metagenome Medicine, Human Genome Center, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
3Division of Innate Immune Regulation, International Research and Development Center for Mucosal Vaccines, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
4Division of Systems Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
5Mucosal Vaccine Project, BIKEN Innovative Vaccine Research Alliance Laboratories, Research Institute for Microbial Diseases, Osaka University, Osaka 565-0871, Japan
6Laboratory of DNA Information Analysis, Human Genome Center, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
7Division of Mucosal Immunology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
8Division of Clinical Genome Research, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
9Department of Computer Science, Tokyo Institute of Technology, Tokyo 152-8550, Japan
10Department of Medicine, University of California San Diego, La Jolla, CA 92093, USA
11Division of Gastroenterology, Department of Medicine, CU-UCSD Center for Mucosal Immunology, Allergy and Vaccines, University of California San Diego, La Jolla, CA 92093, USA
12Division of Comparative Pathology and Medicine, Department of Pathology, University of California San Diego, La Jolla, CA 92093, USA
13Center for Veterinary Sciences and Comparative Medicine, University of California San Diego, La Jolla, CA 92093, USA
14Department of Mucosal Immunology, IMSUT Distinguished Professor Unit, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
15International Research and Development Center for Mucosal Vaccines, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
16Division of Health Medical Data Science, Health Intelligence Center, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
17Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Tokyo 113-8657, Japan
18Lead contact
6Laboratory of DNA Information Analysis, Human Genome Center, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
7Division of Mucosal Immunology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
8Division of Clinical Genome Research, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
9Department of Computer Science, Tokyo Institute of Technology, Tokyo 152-8550, Japan
10Department of Medicine, University of California San Diego, La Jolla, CA 92093, USA
11Division of Gastroenterology, Department of Medicine, CU-UCSD Center for Mucosal Immunology, Allergy and Vaccines, University of California San Diego, La Jolla, CA 92093, USA
12Division of Comparative Pathology and Medicine, Department of Pathology, University of California San Diego, La Jolla, CA 92093, USA
13Center for Veterinary Sciences and Comparative Medicine, University of California San Diego, La Jolla, CA 92093, USA
14Department of Mucosal Immunology, IMSUT Distinguished Professor Unit, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
15International Research and Development Center for Mucosal Vaccines, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
16Division of Health Medical Data Science, Health Intelligence Center, The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan
17Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Tokyo 113-8657, Japan
18Lead contact
DOI:10.1016/j.chom.2020.06.005
Funding
The Takeda Science Foundation, the Canon Foundation, strategic programs for innovative research field 1 from Ministry of Education, Culture, Sports, Science and Technology of Japan, the Center of Innovation Program from Japan Science and Technology Agency.