Research Topics
1. 各種疾患のメカニズム解明に向けた細胞内情報伝達系の大規模数理ネットワーク解析
2. ヒトプロテオームの複雑な全体像を規定する翻訳制御システムの解明
ヒトゲノム配列の解読と並行し、転写産物の網羅的な配列解析も国際レベルで精力的に進められ、約半数の遺伝子が5’末端の非翻訳領域に短いORFを有していることが示されました。翻訳開始システムとして従来提唱されているスキャニングメカニズムに従って、これらの小ORFが実際に細胞中で翻訳されているか否かを実験的に明らかにすることは、ヒトプロテオームの真の全体像を理解するために必要不可欠であると考えられます。私たちは低分子タンパク質に焦点を絞ったプロテオーム解析により、mRNAの5’-端非翻訳領域に由来する翻訳産物を世界で初めて直接同定することに成功しました(Oyama et al., Genome Res., 2004)。また、二次元nanoLC-MS/MSシステムによる大規模な同定解析により、選択的な転写開始やスプライシングによる転写産物の構造調節、並びに翻訳開始部位の調節によって規定される多様なタンパク質コード領域の存在が示され、ヒトプロテオームの多様性に寄与する複雑な翻訳制御システムの一端を明らかにしています(Oyama et al., Mol. Cell. Proteomics, 2007)。大変興味深いことに、従来の想定を超えるこれらの新規タンパク質群の中にリン酸化修飾によって活性制御を受けるシグナル制御因子が存在することも新たに分かってきており(Kozuka-Hata et al., PLoS ONE, 2012)、Orbitrap Eclipse Tribrid質量分析計のReal-Time Searchプラットフォームを活用した高深度定量解析では、リボソームプロファイリングによって規定される新規ORF群に関する大規模な薬剤依存的翻訳制御動態が明らかになっています(Kozuka-Hata et al., Biomolecules, 2023)。
3. 高感度nanoLC-MS/MSシステムを用いた高精度プロテオーム・モディフィコーム・インタラクトーム解析
私たちの研究室では、研究所内外の大学・研究機関と共同で癌・感染症等の多様な疾患や生命現象を対象とする高精度プロテオーム・モディフィコーム・インタラクトーム解析を推進し、従来の分子細胞生物学・生化学的手法では明らかにされてこなかった機能性微量構成因子の同定やリン酸化・ユビキチン化をはじめとする修飾部位に関する精密な一次構造の決定を通して新規知見の導出に幅広く貢献し、多数の共同研究成果を発表しています(Fukui et al., J. Exp. Med., 2009; Arii et al., Nature, 2010; Abe et al., Genes Dev., 2011; Gorai et al., Proc. Natl. Acad. Sci. USA, 2012; Oikawa et al., J. Cell Biol., 2012; Shibata et al., Nat. Commun., 2012; Hirano et al., Cell, 2013; Tokuhara et al., J. Clin. Invest., 2013; Takai et al., Cell Rep., 2014; Ohta et al., Nat. Commun., 2014; Watanabe et al., Cell Host Microbe, 2014; Inoue et al., Leukemia, 2015; Taniue et al., Proc. Natl. Acad. Sci. USA, 2016; Hashimoto et al., Nat. Commun., 2016; Saitoh et al., Nat. Commun., 2017; Inoue et al., Leukemia, 2018; Asada et al., Nat. Commun., 2018; Oishi et al., Cell Rep., 2018; Tatebayashi et al., EMBO J., 2020; Tsuboyama et al., PLoS Biol., 2020; Tanaka et al., PLoS Biol., 2022; Ohe et al., Nat. Commun., 2022; Fujikawa et al., Curr. Biol., 2023; Yoshimoto et al., Mol. Cell, 2023; Kaito et al., Nat. Commun., 2024; Kato et al., Nat. Microbiol., 2025)。
People
Group Member
- 准教授
- 尾山 大明(医学博士)
- 上席技術専門員
- 秦 裕子(生命科学博士)
- 学術/特任専門職員
- 北村 亜矢
- 廣木 朋子
- 相澤 愛子
- 宮村 尚明
- 大学院生
- 全 準浩
- 黃 盛民
- 出向研究員
- 足立 崇
- 客員研究員
- 藤元 次郎
博士号取得者
大学院修士課程修了者
Publications
Publications
Herpes simplex virus 1 evades APOBEC1-mediated immunity via its uracil-DNA glycosylase in mice.
Nat. Microbiol., 10: 1758-1774, 2025.
Masuda S, Kurabayashi N, Nunokawa R, Otobe Y, Kozuka-Hata H, Oyama M, Shibata Y, Inoue JI, Koebis M, Aiba A, Yoshitane H, and Fukada Y.
TRAF7 determines circadian period through ubiquitination and degradation of DBP.
Commun. Biol., 7: 1280, 2024.
Kaito S, Aoyama K, Oshima M, Tsuchiya A, Miyota M, Yamashita M, Koide S, Nakajima-Takagi Y, Kozuka-Hata H, Oyama M, Yogo T, Yabushita T, Ito R, Ueno M, Hirao A, Tohyama K, Li C, Kawabata KC, Yamaguchi K, Furukawa Y, Kosako H, Yoshimi A, Goyama S, Nannya Y, Ogawa S, Agger K, Helin K, Yamazaki S, Koseki H, Doki N, Harada Y, Harada H, Nishiyama A, Nakanishi M, and Iwama A.
Inhibition of TOPORS ubiquitin ligase augments the efficacy of DNA hypomethylating agents through DNMT1 stabilization.
Nat. Commun., 15: 7359, 2024.
Yuki Y, Kurokawa S, Sugiura K, Kashima K, Maruyama S, Yamanoue T, Honma A, Mejima M, Takeyama N, Kuroda M, Kozuka-Hata H, Oyama M, Masumura T, Nakahashi-Ouchida R, Fujihashi K, Hiraizumi T, Goto E, and Kiyono H.
MucoRice-CTB line 19A, a new marker-free transgenic rice-based cholera vaccine produced in an LED-based hydroponic system.
Front. Plant Sci., 15: 1342662, 2024.
Yoshimoto R, Nakayama Y, Nomura I, Yamamoto I, Nakagawa Y, Tanaka S, Kurihara M, Suzuki Y, Kobayashi T, Kozuka-Hata H, Oyama M, Mito M, Iwasaki S, Yamazaki T, Hirose T, Araki K, and Nakagawa S.
4.5SH RNA counteracts deleterious exonization of SINE B1 in mice.
Mol. Cell, 83: 4479-4493.e6, 2023.
Homma MK, Nakato R, Niida A, Bando M, Fujiki K, Yokota N, Yamamoto S, Shibata T, Takagi M, Yamaki J, Kozuka-Hata H, Oyama M, Shirahige K, and Homma Y.
Cell cycle-dependent gene networks for cell proliferation activated by nuclear CK2α complexes.
Life Sci. Alliance, 7: e202302077, 2023.
Soeda S, Oyama M, Kozuka-Hata H, and Yamamoto T.
The CCR4-NOT complex suppresses untimely translational activation of maternal mRNAs.
Development, 150: dev201773, 2023.
Kozuka-Hata H, Hiroki T, Miyamura N, Kitamura A, Tsumoto K, Inoue JI, and Oyama M#.(#: corresponding author)
Real-Time Search-Assisted Multiplexed Quantitative Proteomics Reveals System-Wide Translational Regulation of Non-Canonical Short Open Reading Frames.
Biomolecules, 13: 979, 2023.
Arakawa K, Hirose T, Inada T, Ito T, Kai T, Oyama M, Tomari Y, Yoda T, and Nakagawa S.
Nondomain biopolymers: Flexible molecular strategies to acquire biological functions.
Genes Cells, 28: 539-552, 2023.
Fujikawa D, Nakamura T, Yoshioka D, Li Z, Moriizumi H, Taguchi M, Tokai-Nishizumi N, Kozuka-Hata H, Oyama M, and Takekawa M.
Stress granule formation inhibits stress-induced apoptosis by selectively sequestering executioner caspases.
Curr. Biol., 33: 1967-1981.e8, 2023.
Yamaguchi K, Nakagawa S, Saku A, Isobe Y, Yamaguchi R, Sheridan P, Takane K, Ikenoue T, Zhu C, Miura M, Okawara Y, Nagatoishi S, Kozuka-Hata H, Oyama M, Aikou S, Ahiko Y, Shida D, Tsumoto K, Miyano S, Imoto S, and Furukawa Y.
Bromodomain protein BRD8 regulates cell cycle progression in colorectal cancer cells through a TIP60-independent regulation of the pre-RC complex.
iScience, 26: 106563, 2023.
Miyashita R, Nishiyama A, Qin W, Chiba Y, Kori S, Kato N, Konishi C, Kumamoto S, Kozuka-Hata H, Oyama M, Kawasoe Y, Tsurimoto T, Takahashi TS, Leonhardt H, Arita K, and Nakanishi M.
The termination of UHRF1-dependent PAF15 ubiquitin signaling is regulated by USP7 and ATAD5.
Elife, 12: e79013, 2023.
Ohe S, Kubota Y, Yamaguchi K, Takagi Y, Nashimoto J, Kozuka-Hata H, Oyama M, Furukawa Y, and Takekawa M.
ERK-mediated NELF-A phosphorylation promotes transcription elongation of immediate-early genes by releasing promoter-proximal pausing of RNA polymerase II.
Nat. Commun., 13: 7476, 2022.
Tanaka A, Nakano T, Watanabe K, Masuda K, Honda G, Kamata S, Yasui R, Kozuka-Hata H, Watanabe C, Chinen T, Kitagawa D, Sawai S, Oyama M, Yanagisawa M, and Kunieda T.
Stress-dependent cell stiffening by tardigrade tolerance proteins that reversibly form a filamentous network and gel.
PLoS Biol., 20: e3001780, 2022.
Maeda F, Kato A, Takeshima K, Shibazaki M, Sato R, Shibata T, Miyake K, Kozuka-Hata H, Oyama M, Shimizu E, Imoto S, Miyano S, Adachi S, Natsume T, Takeuchi K, Maruzuru Y, Koyanagi N, Arii J, and Kawaguchi Y.
Role of the Orphan Transporter SLC35E1 in the Nuclear Egress of Herpes Simplex Virus 1.
J. Virol., 96: e0030622, 2022.
Oyama M# and Kozuka-Hata H. (# ; corresponding author)
Integrative Network Analysis of Cancer Cell Signaling by High-Resolution Proteomics.
Springer Proceedings in Mathematics and Statistics, 370: 274-282, 2021.
Sasou A, Yuki Y, Honma A, Sugiura K, Kashima K, Kozuka-Hata H, Nojima M, Oyama M, Kurokawa S, Maruyama S, Kuroda M, Tanoue S, Takamatsu N, Fujihashi K, Goto E, and Kiyono H.
Comparative whole-genome and proteomics analyses of the next seed bank and the original master seed bank of MucoRice-CTB 51A line, a rice-based oral cholera vaccine.
BMC Genomics, 22: 59, 2021.
Watanabe M, Arii J, Takeshima K, Fukui A, Shimojima M, Kozuka-Hata H, Oyama M, Minamitani T, Yasui T, Kubota Y, Takekawa M, Kosugi I, Maruzuru Y, Koyanagi N, Kato A, Mori Y, and Kawaguchi Y.
Prohibitin-1 Contributes to Cell-to-Cell Transmission of Herpes Simplex Virus 1 via the MAPK/ERK Signaling Pathway.
J. Virol., 95: e01413-20, 2021.
Tsuboi Y, Oyama M, Kozuka-Hata H, Ito A, Matsubara D, and Murakami Y.
CADM1 suppresses c-Src activation by binding with Cbp on membrane lipid rafts and intervenes colon carcinogenesis.
Biochem. Biophys. Res. Commun., 529: 854-860, 2020.
Narita K, Nagatomo H, Kozuka-Hata H, Oyama M, and Takeda S.
Discovery of a vertebrate-specific factor that processes flagellar glycolytic enolase during motile ciliogenesis.
iScience, 23: 100992, 2020.
Tsuboyama K, Osaki T, Matsuura-Suzuki E, Kozuka-Hata H, Okada Y, Oyama M, Ikeuchi Y, Iwasaki S, and Tomari Y.
A widespread family of heat-resistant obscure (Hero) proteins protect against protein instability and aggregation.
PLoS Biol., 18: e3000632, 2020.
Kozuka-Hata H#, Kitamura A, Hiroki T, Aizawa A, Tsumoto K, Inoue JI, and Oyama M#.(#: corresponding authors)
System-Wide Analysis of Protein Acetylation and Ubiquitination Reveals a Diversified Regulation in Human Cancer Cells.
Biomolecules, 10: 411, 2020.
Tatebayashi K, Yamamoto K, Tomida T, Nishimura A, Takayama T, Oyama M, Kozuka-Hata H, Adachi-Akahane S, Tokunaga Y, and Saito H.
Osmostress enhances activating phosphorylation of Hog1 MAP kinase by mono-phosphorylated Pbs2 MAP2K.
EMBO J., 39: e103444, 2020.
<2015-2019>
Aoki M, Koga K, Miyazaki M, Hamasaki M, Koshikawa N, Oyama M, Kozuka-Hata H, Seiki M, Toole BP, and Nabeshima K.
CD73 complexes with emmprin to regulate MMP-2 production from co-cultured sarcoma cells and fibroblasts.
BMC Cancer, 19: 912, 2019.
Takano R, Kozuka-Hata H, Kondoh D, Bochimoto H, Oyama M, and Kato K.
A High-Resolution Map of SBP1 Interactomes in Plasmodium falciparum-infected Erythrocytes.
iScience, 19: 703-714, 2019.
Oishi K, Yamayoshi S, Kozuka-Hata H, Oyama M, and Kawaoka Y.
N-Terminal Acetylation by NatB Is Required for the Shutoff Activity of Influenza A Virus PA-X.
Cell Rep., 24: 851-860, 2018.
Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, Yonezawa T, Fujino T, Hayashi Y, Kawabata KC, Fukuyama T, Tanaka Y, Yokoyama A, Yamazaki S, Kozuka-Hata H, Oyama M, Kojima S, Kawazu M, Mano H, and Kitamura T.
Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis.
Nat. Commun., 9: 2733, 2018.
Kato A, Oda S, Watanabe M, Oyama M, Kozuka-Hata H, Koyanagi N, Maruzuru Y, Arii J, and Kawaguchi Y.
Roles of the Phosphorylation of Herpes Simplex Virus 1 UL51 at a Specific Site in Viral Replication and Pathogenicity.
J. Virol., 92: e01035-18, 2018.
Koyanagi N, Kato A, Takeshima K, Maruzuru Y, Kozuka-Hata H, Oyama M, Arii J, and Kawaguchi Y.
Regulation of Herpes Simplex Virus 2 Protein Kinase UL13 by Phosphorylation and Its Role in Viral Pathogenesis.
J. Virol., 92: e00807-18, 2018.
Inoue D, Fujino T, Sheridan P, Zhang YZ, Nagase R, Horikawa S, Li Z, Matsui H, Kanai A, Saika M, Yamaguchi R, Kozuka-Hata H, Kawabata KC, Yokoyama A, Goyama S, Inaba T, Imoto S, Miyano S, Xu M, Yang FC, Oyama M, and Kitamura T.
A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies.
Leukemia, 32: 1327-1337, 2018.
Fujita T*#, Kozuka-Hata H*, Hori Y, Takeuchi J, Kubo T, and Oyama M#.(*; equal contribution) (#: corresponding authors)
Shotgun proteomics deciphered age/division of labor-related functional specification of three honeybee (Apis mellifera L.) exocrine glands.
PLoS One, 13: e0191344, 2018.
Kikuchi K, Kozuka-Hata H, Oyama M, Seiki M, and Koshikawa N.
Identification of Proteolytic Cleavage Sites of EphA2 by Membrane Type 1 Matrix Metalloproteinase on the Surface of Cancer Cells.
Methods Mol. Biol., 1731: 29-37, 2018.
Saitoh SI, Abe F, Kanno A, Tanimura N, Mori Saitoh Y, Fukui R, Shibata T, Sato K, Ichinohe T, Hayashi M, Kubota K, Kozuka-Hata H, Oyama M, Kikko Y, Katada T, Kontani K, and Miyake K.
TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells.
Nat. Commun., 8: 1592, 2017.
Kozuka-Hata H and Oyama M#. (#: corresponding author)
Comprehensive Network Analysis of Cancer Stem Cell Signalling through Systematic Integration of Post-Translational Modification Dynamics.
In Applications of RNA-Seq and Omics Strategies - From Microorganisms to Human Health (ed. Fabio A. Marchi, Priscila D.R. Cirillo and Elvis C. Mateo), InTech, 265-278, 2017.
Contu VR, Hase K, Kozuka-Hata H, Oyama M, Fujiwara Y, Kabuta C, Takahashi M, Hakuno F, Takahashi SI, Wada K, and Kabuta T.
Lysosomal targeting of SIDT2 via multiple YxxΦ motifs is required for SIDT2 function in the process of RNautophagy.
J. Cell Sci., 130: 2843-2853, 2017.
Suzawa M, Noguchi K, Nishi K, Kozuka-Hata H, Oyama M, and Ui-Tei K.
Comprehensive Identification of Nuclear and Cytoplasmic TNRC6A-Associating Proteins.
J. Mol. Biol., 429: 3319-3333, 2017.
Hashimoto T, Horikawa DD, Saito Y, Kuwahara H, Kozuka-Hata H, Shin-I T, Minakuchi Y, Ohishi K, Motoyama A, Aizu T, Enomoto A, Kondo K, Tanaka S, Hara Y, Koshikawa S, Sagara H, Miura T, Yokobori S, Miyagawa K, Suzuki Y, Kubo T, Oyama M, Kohara Y, Fujiyama A, Arakawa K, Katayama T, Toyoda A, and Kunieda T.
Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein.
Nat. Commun., 7: 12808, 2016.
Narushima Y, Kozuka-Hata H, Tsumoto K, Inoue J, and Oyama M#. (#: corresponding author)
Quantitative phosphoproteomics-based molecular network description for high-resolution kinase-substrate interactome analysis.
Bioinformatics, 32: 2083-2088, 2016.
Hirano A, Nakagawa T, Yoshitane H, Oyama M, Kozuka-Hata H, Lanjakornsiripan D, and Fukada Y.
USP7 and TDP-43: Pleiotropic Regulation of Cryptochrome Protein Stability Paces the Oscillation of the Mammalian Circadian Clock.
PLoS One, 11: e0154263, 2016.
Yuki Y, Kurokawa S, Kozuka-Hata H, Tokuhara D, Mejima M, Kuroda M, Oyama M, Nishimaki-Mogami T, Teshima R, and Kiyono H.
Differential analyses of major allergen proteins in wild-type rice and rice producing a fragment of anti-rotavirus antibody.
Regul. Toxicol. Pharmacol., 76: 128-136, 2016.
Nishimura A, Yamamoto K, Oyama M, Kozuka-Hata H, Saito H, and Tatebayashi K.
Scaffold Protein Ahk1, Which Associates with Hkr1, Sho1, Ste11, and Pbs2, Inhibits Cross Talk Signaling from the Hkr1 Osmosensor to the Kss1 Mitogen-Activated Protein Kinase.
Mol. Cell. Biol., 36: 1109-1123, 2016.
Taniue K, Kurimoto A, Sugimasa H, Nasu E, Takeda Y, Iwasaki K, Nagashima T, Okada-Hatakeyama M, Oyama M, Kozuka-Hata H, Hiyoshi M, Kitayama J, Negishi L, Kawasaki Y, and Akiyama T.
Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1.
Proc. Natl. Acad. Sci. USA, 113: 1273-1278, 2016.
Sato Y, Kato A, Maruzuru Y, Oyama M, Kozuka-Hata H, Arii J, and Kawaguchi Y.
Cellular Transcriptional Coactivator RanBP10 and Herpes Simplex Virus 1 ICP0 Interact and Synergistically Promote Viral Gene Expression and Replication.
J. Virol., 90: 3173-3186, 2016.
Narushima Y, Kozuka-Hata H, Koyama-Nasu R, Tsumoto K, Inoue J, Akiyama T, and Oyama M#. (#: corresponding author)
Integrative Network Analysis Combined with Quantitative Phosphoproteomics Reveals TGFBR2 as a Novel Regulator of Glioblastoma Stem Cell Properties.
Mol. Cell. Proteomics, 15: 1017-1031, 2016.
Sato Y, Kato A, Arii J, Koyanagi N, Kozuka-Hata H, Oyama M, and Kawaguchi Y.
Ubiquitin-specific protease 9X in host cells interacts with herpes simplex virus 1 ICP0.
J. Vet. Med. Sci., 78: 405-410, 2016.
Kozuka-Hata H and Oyama M#. (#: corresponding author)
Phosphoproteomics-based network analysis of cancer cell signaling systems.
In Protein Modifications in Pathogenic Dysregulation of Signaling (ed. Jun-ichiro Inoue and Mutsuhiro Takekawa), Springer, 3-15, 2015.
Inoue D, Nishimura K, Kozuka-Hata H, Oyama M, and Kitamura T.
The stability of epigenetic factor ASXL1 is regulated through ubiquitination and USP7-mediated deubiquitination.
Leukemia, 29: 2257-2260, 2015.
Liu Z, Kato A, Oyama M, Kozuka-Hata H, Arii J, and Kawaguchi Y.
Role of Host Cell p32 in Herpes Simplex Virus 1 De-Envelopment during Viral Nuclear Egress.
J. Virol., 89: 8982-8998, 2015.
Hirohata Y, Arii J, Liu Z, Shindo K, Oyama M, Kozuka-Hata H, Sagara H, Kato A, and Kawaguchi Y.
Herpes Simplex Virus 1 Recruits CD98 Heavy Chain and β1 Integrin to the Nuclear Membrane for Viral De-Envelopment.
J. Virol., 89: 7799-7812, 2015.
Kobayashi R, Kato A, Oda S, Koyanagi N, Oyama M, Kozuka-Hata H, Arii J, and Kawaguchi Y.
Function of the Herpes Simplex Virus 1 Small Capsid Protein VP26 Is Regulated by Phosphorylation at a Specific Site.
J. Virol., 89: 6141-6147, 2015.
Hirohata Y, Kato A, Oyama M, Kozuka-Hata H, Koyanagi N, Arii J, and Kawaguchi Y.
Interactome analysis of herpes simplex virus 1 envelope glycoprotein H.
Microbiol. Immunol., 59: 331-337, 2015.
Homma MK, Shibata T, Suzuki T, Ogura M, Kozuka-Hata H, Oyama M, and Homma Y.
Role for protein kinase CK2 on cell proliferation: Assessing CK2 complex components in the nucleus during the cell cycle progression.
In Protein Kinase CK2 Cellular Function in Normal and Disease States (ed. Khalil Ahmed, Olaf-Georg Issinger, and Ryszard Szyszka), Springer, 197-226, 2015.
<2010-2014>
Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, and Kawaoka Y.
Influenza virus-host interactome screen as a platform for antiviral drug development.
Cell Host Microbe, 16: 795-805, 2014.
Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, and Kitagawa D.
Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole.
Nat. Commun., 5: 5267, 2014.
Takai H, Masuda K, Sato T, Sakaguchi Y, Suzuki T, Suzuki T, Koyama-Nasu R, Nasu-Nishimura Y, Katou Y, Ogawa H, Morishita Y, Kozuka-Hata H, Oyama M, Todo T, Ino Y, Mukasa A, Saito N, Toyoshima C, Shirahige K, and Akiyama T.
5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex.
Cell Rep., 9: 48-60, 2014.
Fujii H, Kato A, Mugitani M, Kashima Y, Oyama M, Kozuka-Hata H, Arii J, and Kawaguchi Y.
The UL12 protein of herpes simplex virus 1 is regulated by tyrosine phosphorylation.
J. Virol., 88: 10624-10634, 2014.
Maruzuru Y, Shindo K, Liu Z, Oyama M, Kozuka-Hata H, Arii J, Kato A, and Kawaguchi Y.
Role of herpes simplex virus 1 immediate early protein ICP22 in viral nuclear egress.
J. Virol., 88: 7445-7454, 2014.
Sugai A, Sato H, Hagiwara K, Kozuka-Hata H, Oyama M, Yoneda M, and Kai C.
Newly identified minor phosphorylation site threonine-279 of measles virus nucleoprotein is a prerequisite for nucleocapsid formation.
J. Virol., 88: 1140-1149, 2014.
Kato A, Tsuda S, Liu Z, Kozuka-Hata H, Oyama M, and Kawaguchi Y.
Herpes simplex virus 1 protein kinase Us3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells.
J. Virol., 88: 655-666, 2014.
Sakamoto T, Weng JS, Hara T, Yoshino S, Kozuka-Hata H, Oyama M, and Seiki M.
Hypoxia-inducible factor 1 regulation through cross talk between mTOR and MT1-MMP.
Mol. Cell. Biol., 34: 30-42, 2014.
Yamaguchi N, Oyama M, Kozuka-Hata H, and Inoue J.
Involvement of A20 in the molecular switch that activates the non-canonical NF-кB pathway.
Sci. Rep., 3: 2568, 2013.
Tokuhara D, Alvarez B, Mejima M, Hiroiwa T, Takahashi Y, Kurokawa S, Kuroda M, Oyama M, Kozuka-Hata H, Nochi T, Sagara H, Aladin F, Marcotte H, Frenken LG, Iturriza-Gomara M, Kiyono H, Hammarstrom L, and Yuki Y.
Rice-based oral antibody fragment prophylaxis and therapy against rotavirus infection.
J. Clin. Invest., 123: 3829-3838, 2013.
Maruzuru Y, Fujii H, Oyama M, Kozuka-Hata H, Kato A, and Kawaguchi Y.
Roles of p53 in herpes simplex virus 1 replication.
J. Virol., 87: 9323-9332, 2013.
Kurokawa S, Nakamura R, Mejima M, Kozuka-Hata H, Kuroda M, Takeyama N, Oyama M, Satoh S, Kiyono H, Masumura T, Teshima R, and Yuki Y.
MucoRice-cholera toxin B-subunit, a rice-based oral cholera vaccine, down-regulates the expression of α-amylase/trypsin inhibitor-like protein family as major rice allergens.
J. Proteome Res., 12: 3372-3382, 2013.
Takahashi K, Halfmann P, Oyama M, Kozuka-Hata H, Noda T, and Kawaoka Y.
DNA Topoisomerase 1 Facilitates the Transcription and Replication of the Ebola Virus Genome.
J. Virol., 87: 8862-8869, 2013.
Yuki Y, Mejima M, Kurokawa S, Hiroiwa T, Takahashi Y, Tokuhara D, Nochi T, Katakai Y, Kuroda M, Takeyama N, Kashima K, Abe M, Chen Y, Nakanishi U, Masumura T, Takeuchi Y, Kozuka-Hata H, Shibata H, Oyama M, Tanaka K, and Kiyono H.
Induction of toxin-specific neutralizing immunity by molecularly uniform rice-based oral cholera toxin B subunit vaccine without plant-associated sugar modification.
Plant Biotechnol. J., 11: 799-808, 2013.
Kozuka-Hata H, Goto Y, and Oyama M#. (#: corresponding author)
Phosphoproteomics-based characterization of cancer cell signaling networks.
In Oncogenomics and Cancer Proteomics - Novel Approaches in Biomarkers Discovery and Therapeutic Targets in Cancer (ed. Cesar Lopez-Camarillo and Elena Arechaga-Ocampo), InTech, 185-206, 2013.
Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, and Fukada Y.
FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes.
Cell, 152: 1106-1118, 2013.
Koyama-Nasu R, Takahashi R, Yanagida S, Nasu-Nishimura Y, Oyama M, Kozuka-Hata H, Haruta R, Manabe E, Hoshino-Okubo A, Omi H, Yanaihara N, Okamoto A, Tanaka T, and Akiyama T.
The Cancer Stem Cell Marker CD133 Interacts with Plakoglobin and Controls Desmoglein-2 Protein Levels.
PLoS One, 8: e53710, 2013.
Fujita T, Kozuka-Hata H, Ao-Kondo H, Kunieda T, Oyama M, and Kubo T.
Proteomic analysis of the royal jelly and characterization of the functions of its derivation glands in the honeybee.
J. Proteome Res., 12: 404-411, 2013.
Shibata Y, Oyama M, Kozuka-Hata H, Han X, Tanaka Y, Gohda J, and Inoue J.
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO.
Nat. Commun., 3: 1061, 2012.
Shuo T, Koshikawa N, Hoshino D, Minegishi T, Ao-Kondo H, Oyama M, Sekiya S, Iwamoto S, Tanaka K, and Seiki M.
Detection of the heterogeneous O-glycosylation profile of MT1-MMP expressed in cancer cells by a simple MALDI-MS method.
PLoS One, 7: e43751, 2012.
Kozuka-Hata H, Nasu-Nishimura Y, Koyama-Nasu R, Ao-Kondo H, Tsumoto K, Akiyama T, and Oyama M#. (#: corresponding author)
Phosphoproteome of human glioblastoma initiating cells reveals novel signaling regulators encoded by the transcriptome.
PLoS One, 7: e43398, 2012.
Kozuka-Hata H, Nasu-Nishimura Y, Koyama-Nasu R, Ao-Kondo H, Tsumoto K, Akiyama T, and Oyama M#. (#: corresponding author)
Global proteome analysis of glioblastoma stem cells by high-resolution mass spectrometry.
Current Topics in Peptide & Protein Research, 13: 1-47, 2012.
Narita K, Kozuka-Hata H, Nonami Y, Ao-Kondo H, Suzuki T, Nakamura H, Yamakawa K, Oyama M, Inoue T, and Takeda S.
Proteomic analysis of multiple primary cilia reveals a novel mode of ciliary development in mammals.
Biol. Open, 1: 815-825, 2012.
Oikawa T, Oyama M, Kozuka-Hata H, Uehara S, Udagawa N, Saya H, and Matsuo K.
Tks5-dependent formation of circumferential podosomes/invadopodia mediates cell-cell fusion.
J. Cell Biol., 197: 553-568, 2012.
Abe R, Caaveiro JM, Kozuka-Hata H, Oyama M, and Tsumoto K.
Mapping ultra-weak protein-protein interactions between heme transporters of Staphylococcus aureus.
J. Biol. Chem., 287: 16477-16487, 2012.
Gorai T, Goto H, Noda T, Watanabe T, Kozuka-Hata H, Oyama M, Takano R, Neumann G, Watanabe S, and Kawaoka Y.
F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding.
Proc. Natl. Acad. Sci. U S A, 109: 4615-4620, 2012.
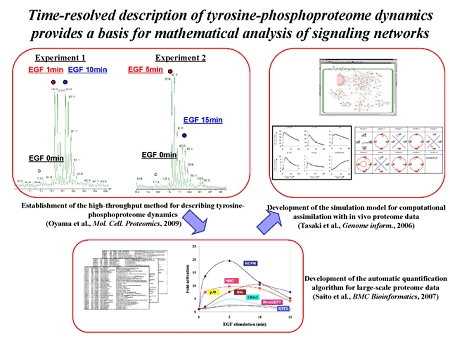
Kozuka-Hata H, Tasaki S, and Oyama M#. (#: corresponding author)
Phosphoproteomics-based systems analysis of signal transduction networks.
Front. Physiol., 2: 113, 2012.
Yamamoto T, Saito M, Kumazawa K, Doi A, Matsui A, Takebe S, Amari T, Oyama M, and Semba K.
ErbB2/HER2: Its Contribution to Basic Cancer Biology and the Development of Molecular Targeted Therapy.
In Breast Cancer - Carcinogenesis, Cell Growth and Signalling Pathways (ed. Mehmet Gunduz and Esra Gunduz), InTech, 139-161, 2011.
Ao-Kondo H, Kozuka-Hata H, and Oyama M#. (#: corresponding author)
Emergence of the Diversified Short ORFeome by Mass Spectrometry-Based Proteomics.
In Computational Biology and Applied Bioinformatics (ed. Heitor Silverio Lopes and Leonardo Magalhaes Cruz), InTech, 417-430, 2011.
Iwasa A, Halfmann P, Noda T, Oyama M, Kozuka-Hata H, Watanabe S, Shimojima M, Watanabe T, and Kawaoka Y.
Contribution of Sec61α to the life cycle of Ebola virus.
J. Infect. Dis., 204: S919-S926, 2011.
Huang M, Sato H, Hagiwara K, Watanabe A, Sugai A, Ikeda F, Kozuka-Hata H, Oyama M, Yoneda M, and Kai C.
Determination of a phosphorylation site in Nipah virus nucleoprotein and its involvement in virus transcription.
J. Gen. Virol., 92: 2133-2141, 2011.
Suzuki M, Kiga K, Kersulyte D, Cok J, Hooper CC, Mimuro H, Sanada T, Suzuki S, Oyama M, Kozuka-Hata H, Kamiya S, Zou QM, Gilman RH, Berg DE, and Sasakawa C.
Attenuated CagA oncoprotein in Helicobacter pylori from Amerindians in Peruvian Amazon.
J. Biol. Chem., 286: 29964-29972, 2011.
Katsumata Y, Kawaguchi Y, Baba S, Hattori S, Tahara K, Ito K, Iwasaki T, Yamaguchi N, Oyama M, Kozuka-Hata H, Hattori H, Nagata K, Yamanaka H, and Hara M.
Identification of three new autoantibodies associated with systemic lupus erythematosus using two proteomic approaches.
Mol. Cell. Proteomics, 10: M110.005330, 2011.
Suzuki T, Kim M, Kozuka-Hata H, Watanabe M, Oyama M, Tsumoto K, and Yamamoto T.
Monoubiquitination of Tob/BTG family proteins competes with degradation-targeting polyubiquitination.
Biochem. Biophys. Res. Commun., 409: 70-74, 2011.
Abe S, Nagasaka K, Hirayama Y, Kozuka-Hata H, Oyama M, Aoyagi Y, Obuse C, and Hirota T.
The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II.
Genes Dev., 25: 863-874, 2011.
Hino N*, Oyama M*, Sato A, Mukai T, Iraha F, Hayashi A, Kozuka-Hata H, Yamamoto T, Yokoyama S, and Sakamoto K. (* ; equal contribution)
Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells.
J. Mol. Biol., 406: 343-353, 2011.
Oyama M*#, Nagashima T*, Suzuki T, Kozuka-Hata H, Yumoto N, Shiraishi Y, Ikeda K, Kuroki Y, Gotoh N, Ishida T, Inoue S, Kitano H, and Okada-Hatakeyama M#. (*; equal contribution) (#: corresponding authors)
Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer.
J. Biol. Chem., 286: 818-829, 2011.
Tasaki S, Nagasaki M, Kozuka-Hata H, Semba K, Gotoh N, Hattori S, Inoue J, Yamamoto T, Miyano S, Sugano S, and Oyama M#. (# ; corresponding author)
Phosphoproteomics-based modeling defines the regulatory mechanism underlying aberrant EGFR signaling.
PLoS One, 5: e13926, 2010.
Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, and Kawaguchi Y.
Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus 1.
Nature, 467: 859-862, 2010.
Matsumura T, Oyama M, Kozuka-Hata H, Ishikawa K, Inoue T, Muta T, Semba K, and Inoue J.
Identification of BCAP-(L) as a negative regulator of the TLR signaling-induced production of IL-6 and IL-10 in macrophages by tyrosine phosphoproteomics.
Biochem. Biophys. Res. Commun., 400: 265-270, 2010.
Fujita T, Kozuka-Hata H, Uno Y, Nishikori K, Morioka M, Oyama M, and Kubo T.
Functional analysis of the honeybee (Apis mellifera L.) salivary system using proteomics.
Biochem. Biophys. Res. Commun., 397: 740-744, 2010.
Oyama M#, Tasaki S, and Kozuka-Hata H. (# ; corresponding author)
Tyrosine-Phosphoproteome Dynamics.
In Systems Biology for Signaling Networks(ed. Choi, S), Springer, 447-454, 2010.
Tsujita K, Itoh T, Kondo A, Oyama M, Kozuka-Hata H, Irino Y, Hasegawa J, and Takenawa T.
Proteome of acidic phospholipid-binding proteins: Spatial and temporal regulation of coronin 1A by phosphoinositides.
J. Biol. Chem., 285: 6781-6789, 2010.
<2005-2009>
Fukui R, Saitoh S, Matsumoto F, Kozuka-Hata H, Oyama M, Tabeta K, Beutler B, and Miyake K.
Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing.
J. Exp. Med., 206: 1339-1350, 2009.
Oyama M#. (# ; corresponding author)
Comprehensive description of signal transduction networks by quantitative proteomics and systems biology.
Curr. Bioinform., 4: 149-153, 2009.
Oyama M*#, Kozuka-Hata H*, Tasaki S, Semba K, Hattori S, Sugano S, Inoue J, and Yamamoto T. (* ; equal contribution) (# ; corresponding author)
Temporal perturbation of tyrosine-phosphoproteome dynamics reveals the system-wide regulatory networks.
Mol. Cell. Proteomics, 8: 226-231, 2009.
Nagashima T*, Oyama M*, Kozuka-Hata H, Yumoto N, Sakaki Y, and Hatakeyama M. (* ; equal contribution)
Phosphoproteome and transcriptome analyses of ErbB ligand-stimulated MCF-7 cells.
Cancer Genomics Proteomics, 5: 161-168, 2008.
Hagiwara K, Sato H, Inoue Y, Watanabe A, Yoneda M, Ikeda F, Fujita K, Fukuda H, Takamura C, Kozuka-Hata H, Oyama M, Sugano S, Ohmi S, and Kai C.
Phosphorylation of measles virus nucleoprotein upregulates the transcriptional activity of minigenomic RNA.
Proteomics, 8: 1871-1879, 2008.
Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, Bjorling E, Blagoev B, Bose R, Brahmachari SK, Burlingame AS, Bustelo XR, Cagney G, Cantin GT, Cardasis HL, Celis JE, Chaerkady R, Chu F, Cole PA, Costello CE, Cotter RJ, Crockett D, DeLany JP, De Marzo AM, DeSouza LV, Deutsch EW, Dransfield E, Drewes G, Droit A, Dunn MJ, Elenitoba-Johnson K, Ewing RM, Van Eyk J, Faca V, Falkner J, Fang X, Fenselau C, Figeys D, Gagne P, Gelfi C, Gevaert K, Gimble JM, Gnad F, Goel R, Gromov P, Hanash SM, Hancock WS, Harsha HC, Hart G, Hays F, He F, Hebbar P, Helsens K, Hermeking H, Hide W, Hjerno K, Hochstrasser DF, Hofmann O, Horn DM, Hruban RH, Ibarrola N, James P, Jensen ON, Jensen PH, Jung P, Kandasamy K, Kheterpal I, Kikuno RF, Korf U, Korner R, Kuster B, Kwon MS, Lee HJ, Lee YJ, Lefevre M, Lehvaslaiho M, Lescuyer P, Levander F, Lim MS, Lobke C, Loo JA, Mann M, Martens L, Martinez-Heredia J, McComb M, McRedmond J, Mehrle A, Menon R, Miller CA, Mischak H, Mohan SS, Mohmood R, Molina H, Moran MF, Morgan JD, Moritz R, Morzel M, Muddiman DC, Nalli A, Navarro JD, Neubert TA, Ohara O, Oliva R, Omenn GS, Oyama M, Paik YK, Pennington K, Pepperkok R, Periaswamy B, Petricoin EF, Poirier GG, Prasad TS, Purvine SO, Rahiman BA, Ramachandran P, Ramachandra YL, Rice RH, Rick J, Ronnholm RH, Salonen J, Sanchez JC, Sayd T, Seshi B, Shankari K, Sheng SJ, Shetty V, Shivakumar K, Simpson RJ, Sirdeshmukh R, Siu KW, Smith JC, Smith RD, States DJ, Sugano S, Sullivan M, Superti-Furga G, Takatalo M, Thongboonkerd V, Trinidad JC, Uhlen M, Vandekerckhove J, Vasilescu J, Veenstra TD, Vidal-Taboada JM, Vihinen M, Wait R, Wang X, Wiemann S, Wu B, Xu T, Yates JR, Zhong J, Zhou M, Zhu Y, Zurbig P, and Pandey A.
Human Proteinpedia enables sharing of human protein data.
Nat. Biotechnol., 26: 164-167, 2008.
Hoshina N, Tezuka T, Yokoyama K, Kozuka-Hata H, Oyama M, and Yamamoto T.
Focal adhesion kinase regulates laminin-induced oligodendroglial process outgrowth.
Genes Cells, 12: 1245-1254, 2007.
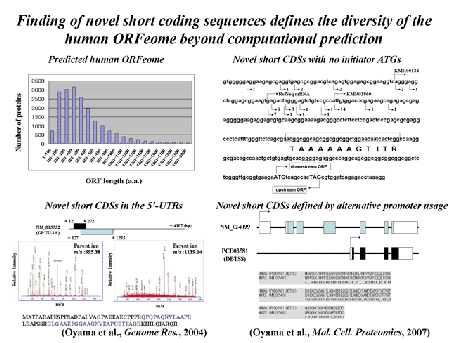
Oyama M#, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, and Sugano S. (# ; corresponding author)
Diversity of translation start sites may define increased complexity of the human short ORFeome.
Mol. Cell. Proteomics, 6: 1000-1006, 2007.
Saito A*, Nagasaki M*, Oyama M*, Kozuka-Hata H, Semba K, Sugano S, Yamamoto T, and Miyano S. (* ; equal contribution)
AYUMS: an algorithm for completely automatic quantitation based on LC-MS/MS proteome data and its application to the analysis of signal transduction.
BMC Bioinformatics, 8: 15, 2007.
Takeda J, Suzuki Y, Nakao M, Barrero RA, Koyanagi KO, Jin L, Motono C, Hata H, Isogai T, Nagai K, Otsuki T, Kuryshev V, Shionyu M, Yura K, Go M, Thierry-Mieg J, Thierry-Mieg D, Wiemann S, Nomura N, Sugano S, Gojobori T, and Imanishi T.
Large-scale identification and characterization of alternative splicing variants of human gene transcripts using 56,419 completely sequenced and manually annotated full-length cDNAs.
Nucleic Acids Res., 34: 3917-3928, 2006.
Tasaki S*, Nagasaki M*, Oyama M, Hata H, Ueno K, Yoshida R, Higuchi T, Sugano S, and Miyano S. (* ; equal contribution)
Modeling and Estimation of Dynamic EGFR Pathway by Data Assimilation Approach using Time Series Proteomic Data.
Genome Inform., 17: 226-238, 2006.
<2000-2004>
Oyama M, Itagaki C, Hata H, Suzuki Y, Izumi T, Natsume T, Isobe T, and Sugano S.
Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs.
Genome Res., 14: 2048-2052, 2004.
Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, and Sugano S.
Complete sequencing and characterization of 21,243 full-length human cDNAs.
Nat. Genet., 36: 40-45, 2004.
Suzuki Y, Taira H, Tsunoda T, Mizushima-Sugano J, Sese J, Hata H, Ota T, Isogai T, Tanaka T, Morishita S, Okubo K, Sakaki Y, Nakamura Y, Suyama A, and Sugano S.
Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites.
EMBO Rep., 2: 388-393, 2001.
Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, Suyama A, Sakaki Y, Morishita S, Okubo K, and Sugano S.
Identification and characterization of the potential promoter regions of 1031 kinds of human genes.
Genome Res., 11: 677-684, 2001.
Oyama M, Ikeda T, Lim T, Ikebukuro K, Masuda Y, and Karube I.
Detection of toxic chemicals with high sensitivity by measuring the quantity of induced P450 mRNAs based on surface plasmon resonance.
Biotechnol. Bioeng., 71: 217-222, 2000-2001.
Lim T, Oyama M, Ikebukuro K, and Karube I.
Detection of atrazine based on the SPR determination of P450 mRNA levels in Saccharomyces cerevisiae.
Anal. Chem., 72: 2856-2860, 2000.
Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, Suyama A, and Sugano S.
Statistical analysis of the 5' untranslated region of human mRNA using "Oligo-Capped" cDNA libraries.
Genomics, 64: 286-297, 2000.
日本語総説、著書
秦 裕子、尾山 大明、井上 純一郎「プロテオミクスの現状と課題―ゲノミクスおよび疾患との関連について―」
医用工学ハンドブック、エヌ・ティー・エス、31-37, 2022.
尾山 大明、秦 裕子「リン酸化プロテオミクスに基づくシグナルネットワーク解析による疾患制御技術の開発」
in silico創薬におけるスクリーニングの高速化・高精度化技術、技術情報協会、473-482, 2018.
尾山 大明、秦 裕子「リン酸化プロテオミクスに基づく数理ネットワーク解析」
実験医学、Vol.35, No.5, 183-186, 2017.
秦 裕子、尾山 大明「ショットガンプロテオミクスが解き明かす高解像度シグナル伝達ネットワーク」
生化学、Vol.85, 456-461, 2013.
尾山 大明、秦 裕子「高精度定量プロテオミクスによる翻訳後修飾シグナルの包括的動態解析」
実験医学、Vol.30, No.5, 681-685, 2012.
尾山 大明「Phos-tagを使った定量プロテオミクスによるリン酸化シグナルネットワークの統合システム解析」
生物物理化学、Vol.56, s29-s32, 2012.
尾山 大明、秦 裕子「リン酸化時系列情報から見えるシグナルダイナミクス」
実験医学 Vol.27, No.16, 2576-2581, 2009.
尾山 大明「翻訳開始点の多様性はヒトの短いORFの複雑な全体像を規定する」
実験医学 Vol.25, No.18, 2895-2898, 2007.
尾山 大明、菅野 純夫「ORF」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、32, 2002.
尾山 大明、菅野 純夫「マルチキャピラリーDNAシークエンサー」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、46, 2002.
尾山 大明、菅野 純夫「マイクロチップ」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、58, 2002.
尾山 大明、菅野 純夫「ナノチップテクノロジー」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、61, 2002.
尾山 大明、菅野 純夫「in silico」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、71, 2002.
尾山 大明、菅野 純夫「クローン」
ゲノム・疾患用語ハンドブック. メディカルレビュー社、178, 2002.
Recruitment & Links
Recruitment
メディカル情報生命専攻
疾患蛋白質工学分野
修士課程・博士課程大学院入学をご希望の方はmoyama[at mark]ims.u-tokyo.ac.jpまでメールにて気軽にご連絡下さい。最先端のプロテオミクス・情報科学技術を用いた次世代の医科学研究・創薬研究に興味がある方の参加を心よりお待ちしています。














 前のページへ
前のページへ