Projects
Analysis of Innate Immune Cells in the Intestinal Lamina Propria
We established the method to isolate lamina propria cells from the small intestine and reported that innate immune cells in the lamina propria are divided into 4 subsets based on the difference in CD11c/CD11b expression patterns:CD11chighCD11blow dendritic cells (DCs), CD11chighCD11bhighDCs, CD11cintCD11bint macrophages (MFs), and CD11cintCD11bhigh eosinophils (Uematsu S, et al. Nat Immunol. 2008.).
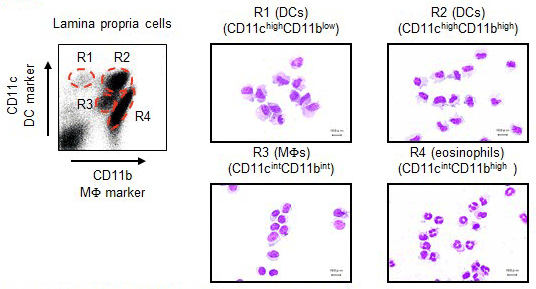
1)Analysis of CD11chighCD11bhigh DCs in the Small Intestinal Lamina Propria
Toll-like receptors (TLRs) recognize distinct microbial components and induce innate immune responses. TLR5 is triggered by bacterial flagellin. Here we generated Tlr5-/- mice and assessed TLR5 function in vivo. Unlike other TLRs, TLR5 was not expressed on conventional dendritic cells or macrophages. In contrast, TLR5 was expressed mainly on intestinal CD11c+ lamina propria cells (LPCs). CD11c+ LPCs detected pathogenic bacteria and secreted proinflammatory cytokines in a TLR5-dependent way. However, CD11c+ LPCs do not express TLR4 and did not secrete proinflammatory cytokines after exposure to a commensal bacterium. Notably, transport of pathogenic Salmonella typhimurium from the intestinal tract to mesenteric lymph nodes was impaired in Tlr5-/- mice. These data suggest that CD11c+ LPCs, via TLR5, detect and are used by pathogenic bacteria in the intestinal lumen (Uematsu S, et al. Nat Immunol. 2006.). We further identify a subset of CD11chighCD11bhigh LPDCs that expressed TLR5 in the small intestine. When stimulated by the TLR5 ligand flagellin, TLR5+ LPDCs induced the differentiation of naive B cells into immunoglobulin (Ig) A-producing plasma cells by a mechanism independent of gut-associated lymphoid tissue. In addition, by a mechanism dependent on TLR5 stimulation, these LPDCs promoted the differentiation of antigen-specific Th17 cells and Th1cells. Unlike spleen DCs, the LPDCs specifically produced retinoic acid, which, in a dose-dependent way, supported the generation and retention of IgA-producing cells in the LP and positively regulated the differentiation Th17 cells. Our findings demonstrate unique properties of LPDCs and the importance of TLR5 for adaptive immunity in the intestine (Uematsu S, et al. Nat Immunol. 2008.).
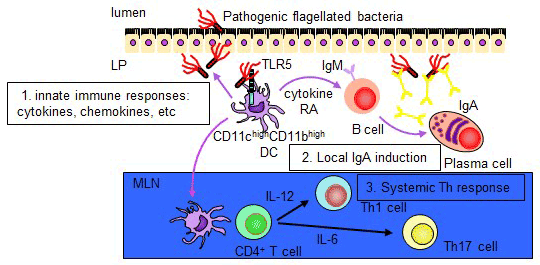
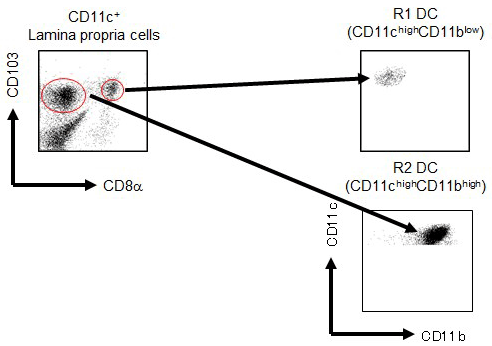
2)Analysis of CD11chighCD11blow DCs in the Small Intestinal Lamina Propria
CD103+ DCs are the major conventional DC population in the intestinal LP. Our previous report showed that low density cells in the LP could be classified into four subsets based on the difference in CD11c/CD11b expression patterns: CD11chighCD11blow DCs, CD11chighCD11bhigh DCs, CD11cintCD11bint MFs, and CD11cintCD11bhigh eosinophils. The CD11chighCD11bhigh DCs, which are CD103+, specifically express TLR5 and induce the differentiation of naive B cells into IgA+ plasma cells. These DCs also mediate the differentiation of Ag-specific Th17 and Th1 cells in response to flagellin. We found that small intestine CD103+ DCs of the LP (LPDCs) could be divided into a small subset of CD8α+ cells and a larger subset of CD8α- cells. Flow cytometry analysis revealed that CD103+CD8α+ and CD103+CD8α- LPDCs were equivalent to CD11chighCD11blow DCs and CD11chighCD11bhigh DCs, respectively. We analyzed a novel subset of CD8α+ LPDCs to elucidate their immunological function. CD103+CD8α+ LPDCs expressed TLR3, TLR7, and TLR9 and produced IL-6 and IL-12p40, but not TNF-α, IL-10, or IL-23, following TLR ligand stimulation. CD103+CD8α+ LPDCs did not express the gene encoding retinoic acid-converting enzyme Raldh2 and were not involved in T cell-independent IgA synthesis or Foxp3+ regulatory T cell induction. Furthermore, CD103+CD8α+ LPDCs induced Ag-specific IgG in serum, a Th1 response, and CTL activity in vivo. Accordingly, CD103+CD8α+ LPDCs exhibit a different function from CD103+CD8α- LPDCs in active immunity. This is the first analysis, to our knowledge, of CD8α+ DCs in the LP of the small intestine (Fujimoto K, et al. J. Immunol. 2011.).
3)Development of Next-generation Mucosal Vaccines
Our laboratory is developing next-generation mucosal vaccines based on the analysis of specific dendritic cells in the lamina propria of the intestinal tract. We discovered mucosal adjuvants that can induce antigen-specific IgA not only in systemic immunity but also in the intestine, despite vaccination by injection. After initial immunization with these mucosal adjuvants, very high titer of antigen-specific secretary IgA (SIgA) can be induced for a long period of time by boosting the oral, respiratory tract and transvaginal antigens only. This immunization method was patented in May 2019 (WO / 2016/199904). This vaccine is capable of inducing a strong mucosal immune response in the mucosa, which is the gate of pathogen invasion, and is expected to develop a completely new concept of "suppressing before onset". In addition, by applying this vaccine method targeting disease-specific intestinal bacteria, it can be used as a new therapeutic approach for various intractable diseases related to disturbance of the intestinal microflora that could not be controlled before (Fujimoto K*, Kawaguchi Y*, Shimohigoshi M*, et al. Gastroenterology. 2019. (*equal contribution)). Currently, this vaccination method is being applied to humans as the next generation mucosal vaccine.

4)Analysis of CD11cintCD11bint MFs and CD11cintCD11bhigh Eosinophils in Small Intestinal Lamina Propria
CD11cintCD11bint MFs express CX3CR1. We are analyzing on the MFs about expression patterns of TLRs, cytokine production, regulatory function and roles in inflammation and infection.
It is well known that large numbers of eosinophils reside in small intestinal lamina propira. However, the function of intestinal eosinophils is yet to be elucidated. We are analyzing on the eosinophils about the activation mechanism and roles in inflammation, parasite infection and food allergy.
Analysis of the Role of Innate Immunity in Radiation Syndrome
1)Role of TLR3 in Acute Radiation Syndrome
High-dose ionizing radiation induces severe DNA damage in the epithelial stem cells in small intestinal crypts and causes gastrointestinal syndrome (GIS). Although the tumour suppressor p53 is a primary factor inducing death of crypt cells with DNA damage, its essential role in maintaining genome stability means inhibiting p53 to prevent GIS is not a viable strategy. Here we show that the innate immune receptor Toll-like receptor 3 (TLR3) is critical for the pathogenesis of GIS. Tlr3−/− mice show substantial resistance to GIS owing to significantly reduced radiation-induced crypt cell death. Despite showing reduced crypt cell death, p53-dependent crypt cell death is not impaired in Tlr3−/− mice. p53-dependent crypt cell death causes leakage of cellular RNA, which induces extensive cell death via TLR3. An inhibitor of TLR3–RNA binding ameliorates GIS by reducing crypt cell death. Thus, we propose blocking TLR3 activation as a novel approach to treat GIS (Takemura N, et al. Nat Commun. 2014.).
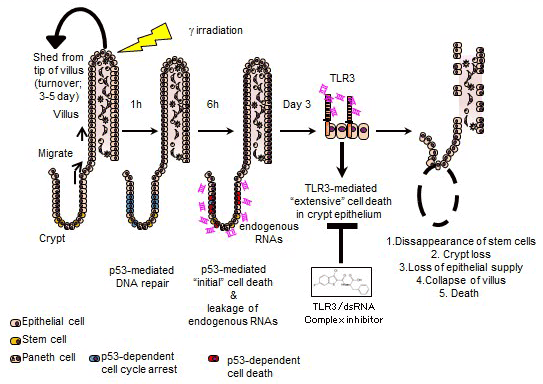
2)Development of New Therapies for Radiation-induced Fibrosis
Radiation-induced intestinal fibrosis is a serious complication after abdominal radiation therapy for pelvic tumors or peritoneal metastases, causing significant fibrosis, especially in the submucosa of the small intestine. This condition was associated with excessive infiltration of activated eosinophils in the submucosa. After irradiation of the abdomen, chronic cell death was induced in the intestinal crypt, causing adenosine triphosphate (ATP) leakage outside the cell and activating myofibroblasts directly under the crypt. This activated myofibroblast was found to infiltrate and activate eosinophils under the mucous membrane. Activated eosinophils under the mucus membrane have been shown to stimulate collagen production from myofibroblasts through the production of TGF-β and induce significant fibrosis under the mucosa. In collaboration with Kyowa Hakko Kirin Co., Ltd., we have developed a new eosinophil-removing antibody targeting mouse interleukin (IL) -5 receptor (R) α. This antibody abolished intestinal eosinophil infiltration and markedly improved radiation-induced intestinal fibrosis. Based on the above, we were able to demonstrate a new treatment strategy for radiation-induced intestinal fibrosis by performing antibody treatment targeting eosinophils (Takemura N, et al. Sci Trans Med. 2018.). We believe that administration of eosinophil-removing antibody can suppress fibrosis of the intestinal tract, prevent complications in patients after radiation therapy, and avoid a decrease in quality of life (QOL).
Metagenome Analysis
1)Development of Database for Intestinal Bacteriomes and Viromes in Human
We established super-high-speed analysis pipeline of intestinal bacteriomes and viromes in collaboration with Prof. Seiya Imoto of the Health Intelligence Center of the University of Tokyo and Prof. Satoru Miyano of the Human Genome Center using the supercomputer Shirokane4 and Shirokane5 of the Institute of Medical Science. Using this pipeline, we are analyzing feces of healthy Japanese individuals and are constructing the world's first database of intestinal bacteriomes and viromes in the same feces. We are now proceeding with host identification of bacteriophages from sequence data. We are building a foundation for new bacteriophage therapy that can control intestinal pathogenic bacteria.
2)Development of New Therapies for Dysbiosis-related Disorders Based on Metagenomics
Recently, dysbiosis has been shown to be involved not only in intestinal tract diseases such as inflammatory bowel disease, but also in autoimmune diseases, diabetes, cardiovascular diseases, autism, etc., and it is clear that they are closely related to the disease onset and activity. Our laboratory conducts metagenomic analysis of intestinal bacteria and virus flora in dysbiosis-related diseases, and is working on the search for microorganisms related to pathological conditions and the development of new therapies.
